Training a GAP model
steps
1. generate a small dataset of water structures
- use CP2K if you havea access to it
- otherwise: use any simple potential implemented in ASE, just for trying this out I have used EMT here
1. generate e0 values
1. separate a training and a validation dataset
1. **train the model**
1. look at the outcome of the model
here we will fit twice, to see the difference between a 2b-only and a 2b+3b fit
[1]:
# general imports
import numpy as np
import matplotlib.pyplot as plt
from copy import deepcopy as cp
# ase imports
import ase.io
from ase import Atoms, Atom
from ase import units
from ase.build import molecule
# for MD
from ase.md.langevin import Langevin
from ase.io.trajectory import Trajectory
[2]:
# helper functions
def make_water(density, super_cell=[3, 3, 3]):
""" Geenrates a supercell of water molecules with a desired density.
Density in g/cm^3!!!"""
h2o = molecule('H2O')
a = np.cbrt((sum(h2o.get_masses()) * units.m ** 3 * 1E-6 ) / (density * units.mol))
h2o.set_cell((a, a, a))
h2o.set_pbc((True, True, True))
#return cp(h2o.repeat(super_cell))
return h2o.repeat(super_cell)
def rms_dict(x_ref, x_pred):
""" Takes two datasets of the same shape and returns a dictionary containing RMS error data"""
x_ref = np.array(x_ref)
x_pred = np.array(x_pred)
if np.shape(x_pred) != np.shape(x_ref):
raise ValueError('WARNING: not matching shapes in rms')
error_2 = (x_ref - x_pred) ** 2
average = np.sqrt(np.average(error_2))
std_ = np.sqrt(np.var(error_2))
return {'rmse': average, 'std': std_}
generating data only with ASE, using the EMT calculator
This is only for the demonstration of how to do it, this run is will be done very fast. There is no practical use of the data beyond lerning the use teach_sparse, quip, etc. with it.
[3]:
# Running MD with ASE's EMT
from ase.calculators.emt import EMT
calc = EMT()
T = 150 # Kelvin
# Set up a grid of water
water = make_water(1.0, [3, 3, 3])
water.set_calculator(calc)
# We want to run MD using the Langevin algorithm
# with a time step of 1 fs, the temperature T and the friction
# coefficient to 0.002 atomic units.
dyn = Langevin(water, 1 * units.fs, T * units.kB, 0.0002)
def printenergy(a=water): # store a reference to atoms in the definition.
"""Function to print the potential, kinetic and total energy."""
epot = a.get_potential_energy() / len(a)
ekin = a.get_kinetic_energy() / len(a)
print('Energy per atom: Epot = %.3feV Ekin = %.3feV (T=%3.0fK) '
'Etot = %.3feV' % (epot, ekin, ekin / (1.5 * units.kB), epot + ekin))
dyn.attach(printenergy, interval=5)
# We also want to save the positions of all atoms after every 5th time step.
traj = Trajectory('dyn_emt.traj', 'w', water)
dyn.attach(traj.write, interval=5)
# Now run the dynamics
printenergy(water)
dyn.run(600) # CHANGE THIS IF YOU WANT LONGER/SHORTER RUN
Energy per atom: Epot = 0.885eV Ekin = 0.000eV (T= 0K) Etot = 0.885eV
Energy per atom: Epot = 0.885eV Ekin = 0.000eV (T= 0K) Etot = 0.885eV
Energy per atom: Epot = 0.820eV Ekin = 0.053eV (T=413K) Etot = 0.874eV
Energy per atom: Epot = 0.660eV Ekin = 0.208eV (T=1610K) Etot = 0.868eV
Energy per atom: Epot = 0.632eV Ekin = 0.224eV (T=1733K) Etot = 0.856eV
Energy per atom: Epot = 0.869eV Ekin = 0.005eV (T= 39K) Etot = 0.874eV
Energy per atom: Epot = 0.781eV Ekin = 0.088eV (T=683K) Etot = 0.869eV
Energy per atom: Epot = 0.756eV Ekin = 0.117eV (T=904K) Etot = 0.873eV
Energy per atom: Epot = 0.706eV Ekin = 0.165eV (T=1274K) Etot = 0.870eV
Energy per atom: Epot = 0.703eV Ekin = 0.159eV (T=1227K) Etot = 0.861eV
Energy per atom: Epot = 0.869eV Ekin = 0.005eV (T= 40K) Etot = 0.874eV
Energy per atom: Epot = 0.693eV Ekin = 0.171eV (T=1323K) Etot = 0.864eV
Energy per atom: Epot = 0.670eV Ekin = 0.198eV (T=1529K) Etot = 0.868eV
Energy per atom: Epot = 0.821eV Ekin = 0.052eV (T=402K) Etot = 0.873eV
Energy per atom: Epot = 0.775eV Ekin = 0.093eV (T=721K) Etot = 0.869eV
Energy per atom: Epot = 0.804eV Ekin = 0.068eV (T=524K) Etot = 0.872eV
Energy per atom: Epot = 0.745eV Ekin = 0.126eV (T=975K) Etot = 0.871eV
Energy per atom: Epot = 0.740eV Ekin = 0.129eV (T=996K) Etot = 0.869eV
Energy per atom: Epot = 0.779eV Ekin = 0.093eV (T=721K) Etot = 0.872eV
Energy per atom: Epot = 0.780eV Ekin = 0.094eV (T=728K) Etot = 0.874eV
Energy per atom: Epot = 0.746eV Ekin = 0.125eV (T=967K) Etot = 0.871eV
Energy per atom: Epot = 0.743eV Ekin = 0.127eV (T=980K) Etot = 0.870eV
Energy per atom: Epot = 0.720eV Ekin = 0.149eV (T=1153K) Etot = 0.869eV
Energy per atom: Epot = 0.745eV Ekin = 0.126eV (T=975K) Etot = 0.871eV
Energy per atom: Epot = 0.759eV Ekin = 0.113eV (T=875K) Etot = 0.872eV
Energy per atom: Epot = 0.726eV Ekin = 0.144eV (T=1114K) Etot = 0.870eV
Energy per atom: Epot = 0.722eV Ekin = 0.148eV (T=1142K) Etot = 0.869eV
Energy per atom: Epot = 0.742eV Ekin = 0.128eV (T=993K) Etot = 0.871eV
Energy per atom: Epot = 0.752eV Ekin = 0.120eV (T=931K) Etot = 0.872eV
Energy per atom: Epot = 0.725eV Ekin = 0.146eV (T=1129K) Etot = 0.871eV
Energy per atom: Epot = 0.718eV Ekin = 0.153eV (T=1184K) Etot = 0.871eV
Energy per atom: Epot = 0.686eV Ekin = 0.184eV (T=1423K) Etot = 0.870eV
Energy per atom: Epot = 0.662eV Ekin = 0.207eV (T=1605K) Etot = 0.869eV
Energy per atom: Epot = 0.636eV Ekin = 0.233eV (T=1801K) Etot = 0.869eV
Energy per atom: Epot = 0.619eV Ekin = 0.251eV (T=1941K) Etot = 0.869eV
Energy per atom: Epot = 0.583eV Ekin = 0.286eV (T=2215K) Etot = 0.869eV
Energy per atom: Epot = 0.606eV Ekin = 0.265eV (T=2048K) Etot = 0.870eV
Energy per atom: Epot = 0.557eV Ekin = 0.312eV (T=2415K) Etot = 0.869eV
Energy per atom: Epot = 0.557eV Ekin = 0.311eV (T=2405K) Etot = 0.868eV
Energy per atom: Epot = 0.581eV Ekin = 0.288eV (T=2228K) Etot = 0.869eV
Energy per atom: Epot = 0.629eV Ekin = 0.241eV (T=1868K) Etot = 0.870eV
Energy per atom: Epot = 0.595eV Ekin = 0.276eV (T=2134K) Etot = 0.870eV
Energy per atom: Epot = 0.580eV Ekin = 0.290eV (T=2247K) Etot = 0.870eV
Energy per atom: Epot = 0.608eV Ekin = 0.262eV (T=2029K) Etot = 0.870eV
Energy per atom: Epot = 0.542eV Ekin = 0.326eV (T=2525K) Etot = 0.869eV
Energy per atom: Epot = 0.519eV Ekin = 0.350eV (T=2706K) Etot = 0.869eV
Energy per atom: Epot = 0.553eV Ekin = 0.318eV (T=2457K) Etot = 0.871eV
Energy per atom: Epot = 0.553eV Ekin = 0.317eV (T=2455K) Etot = 0.870eV
Energy per atom: Epot = 0.563eV Ekin = 0.307eV (T=2377K) Etot = 0.870eV
Energy per atom: Epot = 0.555eV Ekin = 0.315eV (T=2438K) Etot = 0.870eV
Energy per atom: Epot = 0.535eV Ekin = 0.336eV (T=2599K) Etot = 0.871eV
Energy per atom: Epot = 0.522eV Ekin = 0.348eV (T=2693K) Etot = 0.870eV
Energy per atom: Epot = 0.544eV Ekin = 0.326eV (T=2524K) Etot = 0.870eV
Energy per atom: Epot = 0.480eV Ekin = 0.391eV (T=3022K) Etot = 0.871eV
Energy per atom: Epot = 0.487eV Ekin = 0.385eV (T=2979K) Etot = 0.872eV
Energy per atom: Epot = 0.486eV Ekin = 0.384eV (T=2971K) Etot = 0.870eV
Energy per atom: Epot = 0.498eV Ekin = 0.371eV (T=2871K) Etot = 0.870eV
Energy per atom: Epot = 0.483eV Ekin = 0.388eV (T=2998K) Etot = 0.870eV
Energy per atom: Epot = 0.487eV Ekin = 0.382eV (T=2957K) Etot = 0.869eV
Energy per atom: Epot = 0.493eV Ekin = 0.377eV (T=2916K) Etot = 0.870eV
Energy per atom: Epot = 0.482eV Ekin = 0.388eV (T=3005K) Etot = 0.870eV
Energy per atom: Epot = 0.461eV Ekin = 0.410eV (T=3172K) Etot = 0.871eV
Energy per atom: Epot = 0.480eV Ekin = 0.390eV (T=3021K) Etot = 0.871eV
Energy per atom: Epot = 0.449eV Ekin = 0.423eV (T=3275K) Etot = 0.872eV
Energy per atom: Epot = 0.465eV Ekin = 0.406eV (T=3140K) Etot = 0.871eV
Energy per atom: Epot = 0.431eV Ekin = 0.440eV (T=3406K) Etot = 0.871eV
Energy per atom: Epot = 0.393eV Ekin = 0.478eV (T=3695K) Etot = 0.870eV
Energy per atom: Epot = 0.424eV Ekin = 0.448eV (T=3464K) Etot = 0.871eV
Energy per atom: Epot = 0.396eV Ekin = 0.475eV (T=3678K) Etot = 0.871eV
Energy per atom: Epot = 0.389eV Ekin = 0.482eV (T=3728K) Etot = 0.871eV
Energy per atom: Epot = 0.383eV Ekin = 0.488eV (T=3773K) Etot = 0.870eV
Energy per atom: Epot = 0.440eV Ekin = 0.432eV (T=3343K) Etot = 0.872eV
Energy per atom: Epot = 0.416eV Ekin = 0.455eV (T=3517K) Etot = 0.871eV
Energy per atom: Epot = 0.449eV Ekin = 0.424eV (T=3283K) Etot = 0.873eV
Energy per atom: Epot = 0.461eV Ekin = 0.411eV (T=3182K) Etot = 0.872eV
Energy per atom: Epot = 0.499eV Ekin = 0.374eV (T=2895K) Etot = 0.873eV
Energy per atom: Epot = 0.441eV Ekin = 0.430eV (T=3326K) Etot = 0.870eV
Energy per atom: Epot = 0.425eV Ekin = 0.444eV (T=3434K) Etot = 0.868eV
Energy per atom: Epot = 0.423eV Ekin = 0.448eV (T=3464K) Etot = 0.871eV
Energy per atom: Epot = 0.436eV Ekin = 0.435eV (T=3367K) Etot = 0.871eV
Energy per atom: Epot = 0.396eV Ekin = 0.473eV (T=3662K) Etot = 0.869eV
Energy per atom: Epot = 0.395eV Ekin = 0.473eV (T=3657K) Etot = 0.868eV
Energy per atom: Epot = 0.375eV Ekin = 0.493eV (T=3812K) Etot = 0.868eV
Energy per atom: Epot = 0.348eV Ekin = 0.518eV (T=4009K) Etot = 0.867eV
Energy per atom: Epot = 0.361eV Ekin = 0.507eV (T=3922K) Etot = 0.868eV
Energy per atom: Epot = 0.389eV Ekin = 0.479eV (T=3709K) Etot = 0.869eV
Energy per atom: Epot = 0.369eV Ekin = 0.491eV (T=3795K) Etot = 0.859eV
Energy per atom: Epot = 0.357eV Ekin = 0.512eV (T=3957K) Etot = 0.868eV
Energy per atom: Epot = 0.365eV Ekin = 0.504eV (T=3903K) Etot = 0.870eV
Energy per atom: Epot = 0.392eV Ekin = 0.495eV (T=3832K) Etot = 0.887eV
Energy per atom: Epot = 0.390eV Ekin = 0.493eV (T=3817K) Etot = 0.884eV
Energy per atom: Epot = 0.439eV Ekin = 0.487eV (T=3771K) Etot = 0.927eV
Energy per atom: Epot = 0.406eV Ekin = 0.520eV (T=4020K) Etot = 0.926eV
Energy per atom: Epot = 0.378eV Ekin = 0.547eV (T=4231K) Etot = 0.925eV
Energy per atom: Epot = 0.359eV Ekin = 0.566eV (T=4380K) Etot = 0.925eV
Energy per atom: Epot = 0.411eV Ekin = 0.515eV (T=3985K) Etot = 0.926eV
Energy per atom: Epot = 0.361eV Ekin = 0.572eV (T=4426K) Etot = 0.934eV
Energy per atom: Epot = 0.365eV Ekin = 0.566eV (T=4379K) Etot = 0.931eV
Energy per atom: Epot = 0.414eV Ekin = 0.522eV (T=4042K) Etot = 0.937eV
Energy per atom: Epot = 0.417eV Ekin = 0.518eV (T=4010K) Etot = 0.935eV
Energy per atom: Epot = 0.354eV Ekin = 0.574eV (T=4443K) Etot = 0.928eV
Energy per atom: Epot = 0.366eV Ekin = 0.564eV (T=4361K) Etot = 0.930eV
Energy per atom: Epot = 0.394eV Ekin = 0.536eV (T=4145K) Etot = 0.930eV
Energy per atom: Epot = 0.393eV Ekin = 0.541eV (T=4183K) Etot = 0.933eV
Energy per atom: Epot = 0.383eV Ekin = 0.541eV (T=4189K) Etot = 0.924eV
Energy per atom: Epot = 0.426eV Ekin = 0.504eV (T=3901K) Etot = 0.930eV
Energy per atom: Epot = 0.374eV Ekin = 0.555eV (T=4294K) Etot = 0.929eV
Energy per atom: Epot = 0.393eV Ekin = 0.536eV (T=4144K) Etot = 0.928eV
Energy per atom: Epot = 0.390eV Ekin = 0.539eV (T=4173K) Etot = 0.929eV
Energy per atom: Epot = 0.374eV Ekin = 0.554eV (T=4285K) Etot = 0.928eV
Energy per atom: Epot = 0.349eV Ekin = 0.578eV (T=4470K) Etot = 0.927eV
Energy per atom: Epot = 0.409eV Ekin = 0.521eV (T=4034K) Etot = 0.931eV
Energy per atom: Epot = 0.370eV Ekin = 0.560eV (T=4335K) Etot = 0.930eV
Energy per atom: Epot = 0.432eV Ekin = 0.517eV (T=4003K) Etot = 0.949eV
Energy per atom: Epot = 0.435eV Ekin = 0.522eV (T=4038K) Etot = 0.957eV
Energy per atom: Epot = 0.428eV Ekin = 0.534eV (T=4133K) Etot = 0.962eV
Energy per atom: Epot = 0.378eV Ekin = 0.587eV (T=4541K) Etot = 0.965eV
Energy per atom: Epot = 0.397eV Ekin = 0.566eV (T=4381K) Etot = 0.963eV
Energy per atom: Epot = 0.379eV Ekin = 0.581eV (T=4496K) Etot = 0.960eV
Energy per atom: Epot = 0.353eV Ekin = 0.602eV (T=4659K) Etot = 0.955eV
Energy per atom: Epot = 0.425eV Ekin = 0.533eV (T=4124K) Etot = 0.958eV
Energy per atom: Epot = 0.402eV Ekin = 0.571eV (T=4417K) Etot = 0.973eV
[3]:
True
[4]:
# wrap and save traj in .xyz --- the .traj is a non human readable database file, xyz is much better
out_traj = ase.io.read('dyn_emt.traj', ':')
for at in out_traj:
at.wrap()
if 'momenta' in at.arrays: del at.arrays['momenta']
ase.io.write('dyn_emt.xyz', out_traj, 'xyz')
get e0 for H and O - energies of the isolated atoms
This is the energy of the isolated atom, will be in the teach_sparse string in the following format: e0={H:energy:O:energy}
[5]:
isolated_H = Atoms('H', calculator=EMT(), cell=[20, 20, 20], pbc=True)
isolated_O = Atoms('O', calculator=EMT(), cell=[20, 20, 20], pbc=True)
print('e0_H:',isolated_H.get_potential_energy())
print('e0_O:',isolated_O.get_potential_energy())
# this made the e0 string be the following: e0={H:3.21:O:4.6}
e0_H: 3.21
e0_O: 4.6
separate the dataset into a training and a validation set
As we have 120 frames from the 600fs MD, I will do it 60,60 with taking even and odd frames for the two
[6]:
ase.io.write('train.xyz', out_traj[0::2] + [isolated_H] + [isolated_O])
ase.io.write('validate.xyz', out_traj[1::2])
[7]:
out_traj[0].arrays.keys()
[7]:
dict_keys(['numbers', 'positions'])
train our GAP model from the command line
Will use a fit of 2b only, using the desciptor distance_2b.
Let’s understand how this works. The bash command takes named arguments separated by spaces.
teach_sparsethe command which actually does the fite0={H:3.21:O:4.6}the energies of the isolated atomsenergy_parameter_name=energy force_parameter_name=forcesnames of the parametersdo_copy_at_file=F sparse_separate_file=Tjust needed, don’t want to copy the training data and using separate files for the xml makes it fastergp_file=GAP.xmlfilename of the potential parameters, I have always used this name, because I had separate directories for the different trainings potentialsat_file=train.xyztraining filedefault_sigma={0.008 0.04 0 0}sigma values to be used for energies, forces, stresses, hessians in order; this represents the accuracy of the data and the relative weight of them in the fit (more accurate –> more significant in the fit)gap={...}the potential to be fit, separated by ‘:’
distance_2b - cutoff=4.0 radial, practically the highest distance the descriptor takes into account - covariance_type=ard_se use gausses in the fit - delta=0.5 what relative portion of the things shall be determined by this potential - theta_uniform=1.0 width of the gaussians - sparse_method=uniform use uniform bins to choose the sparse points - add_species=T take the species into account, so it will generate more GAPs automatically (see the output) - n_sparse=10
number of sparse points
notice, that the script is running in parallel, using all 8 cores of the current machine
[12]:
! gap_fit energy_parameter_name=energy force_parameter_name=forces do_copy_at_file=F sparse_separate_file=T gp_file=GAP.xml at_file=train.xyz default_sigma={0.008 0.04 0 0} gap={distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform add_species=T n_sparse=10}
libAtoms::Hello World: 07/10/2019 14:37:12
libAtoms::Hello World: git version git@github.com:libAtoms/QUIP.git,f538cd9fe-dirty
libAtoms::Hello World: QUIP_ARCH linux_x86_64_gfortran_openmp
libAtoms::Hello World: compiled on Oct 1 2019 at 21:54:22
libAtoms::Hello World: OpenMP parallelisation with 4 threads
WARNING: libAtoms::Hello World: environment variable OMP_STACKSIZE not set explicitly. The default value - system and compiler dependent - may be too small for some applications.
libAtoms::Hello World: Random Seed = 52632114
libAtoms::Hello World: global verbosity = 0
Calls to system_timer will do nothing by default
================================ Input parameters ==============================
at_file = train.xyz
gap = "distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform add_species=T n_sparse=10"
e0 = 0.0
local_property0 = 0.0
e0_offset = 0.0
e0_method = isolated
default_sigma = "0.008 0.04 0 0"
sparse_jitter = 1.0e-10
hessian_delta = 1.0e-2
core_param_file = quip_params.xml
core_ip_args =
energy_parameter_name = energy
local_property_parameter_name = local_property
force_parameter_name = forces
virial_parameter_name = virial
hessian_parameter_name = hessian
config_type_parameter_name = config_type
sigma_parameter_name = sigma
config_type_sigma =
sigma_per_atom = T
do_copy_at_file = F
sparse_separate_file = T
sparse_use_actual_gpcov = F
gp_file = GAP.xml
verbosity = NORMAL
rnd_seed = -1
do_ip_timing = F
template_file = template.xyz
sparsify_only_no_fit = F
======================================== ======================================
============== Gaussian Approximation Potentials - Database fitting ============
Initial parsing of command line arguments finished.
Found 1 GAPs.
Descriptors have been parsed
XYZ file read
Old GAP: {distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform add_species=T n_sparse=10}
New GAP: {distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=8 Z2=8}
New GAP: {distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=8 Z2=1}
New GAP: {distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=1 Z2=1}
Multispecies support added where requested
===================== Report on number of descriptors found ====================
---------------------------------------------------------------------
Descriptor: distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=8 Z2=8
Number of descriptors: 13192
Number of partial derivatives of descriptors: 79152
---------------------------------------------------------------------
Descriptor: distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=8 Z2=1
Number of descriptors: 63476
Number of partial derivatives of descriptors: 380856
---------------------------------------------------------------------
Descriptor: distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=1 Z2=1
Number of descriptors: 58986
Number of partial derivatives of descriptors: 353916
======================================== ======================================
E0/atom = 0.32100000000000000E+001 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.45999999999999996E+001 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000
========== Report on number of target properties found in training XYZ: ========
Number of target energies (property name: energy) found: 63
Number of target local_properties (property name: local_property) found: 0
Number of target forces (property name: forces) found: 14829
Number of target virials (property name: virial) found: 0
Number of target Hessian eigenvalues (property name: hessian) found: 0
================================= End of report ================================
===== Report on per-configuration/per-atom sigma (error parameter) settings ====
Number of per-configuration setting of energy_sigma found: 0
Number of per-configuration setting of force_sigma found: 0
Number of per-configuration setting of virial_sigma found: 0
Number of per-configuration setting of hessian_sigma found: 0
Number of per-atom setting of force_atom_sigma found: 0
================================= End of report ================================
Cartesian coordinates transformed to descriptors
Started sparse covariance matrix calculation of coordinate 1
Finished sparse covariance matrix calculation of coordinate 1
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1_sparse done in .12079600000000001 cpu secs, .33371098999850801E-001 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1 done in .12085800000000002 cpu secs, .33404888999939431E-001 wall clock secs.
Started sparse covariance matrix calculation of coordinate 2
Finished sparse covariance matrix calculation of coordinate 2
TIMER: gpFull_covarianceMatrix_sparse_Coordinate2_sparse done in .51746400000000004 cpu secs, .13925121800002671 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate2 done in .51753099999999996 cpu secs, .13928714700068667 wall clock secs.
Started sparse covariance matrix calculation of coordinate 3
Finished sparse covariance matrix calculation of coordinate 3
TIMER: gpFull_covarianceMatrix_sparse_Coordinate3_sparse done in .47832100000000000 cpu secs, .12939972799995303 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate3 done in .47839199999999993 cpu secs, .12943785000061325 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_LinearAlgebra done in .58676999999999868E-001 cpu secs, .17518616999950609E-001 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_FunctionValues done in .20999999999826713E-004 cpu secs, .21754999579570722E-004 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse done in 1.1780140000000001 cpu secs, .32216350199996668 wall clock secs.
TIMER: GP sparsify done in 1.2460960000000001 cpu secs, .35651906000020972 wall clock secs.
libAtoms::Finalise: 07/10/2019 14:37:12
libAtoms::Finalise: Bye-Bye!
use the potential with QUIP on trani.xyz and validate.xyz
[17]:
# calculate train.xyz
! quip E=T F=T atoms_filename=train.xyz param_filename=GAP.xml | grep AT | sed 's/AT//' > quip_train.xyz
! quip E=T F=T atoms_filename=validate.xyz param_filename=GAP.xml | grep AT | sed 's/AT//' > quip_validate.xyz
make simple plots of the energies and forces on the EMT and GAP datas
[18]:
def energy_plot(in_file, out_file, ax, title='Plot of energy'):
""" Plots the distribution of energy per atom on the output vs the input"""
# read files
in_atoms = ase.io.read(in_file, ':')
out_atoms = ase.io.read(out_file, ':')
# list energies
ener_in = [at.get_potential_energy() / len(at.get_chemical_symbols()) for at in in_atoms]
ener_out = [at.get_potential_energy() / len(at.get_chemical_symbols()) for at in out_atoms]
# scatter plot of the data
ax.scatter(ener_in, ener_out)
# get the appropriate limits for the plot
for_limits = np.array(ener_in +ener_out)
elim = (for_limits.min() - 0.05, for_limits.max() + 0.05)
ax.set_xlim(elim)
ax.set_ylim(elim)
# add line of slope 1 for refrence
ax.plot(elim, elim, c='k')
# set labels
ax.set_ylabel('energy by GAP / eV')
ax.set_xlabel('energy by EMT / eV')
#set title
ax.set_title(title)
# add text about RMSE
_rms = rms_dict(ener_in, ener_out)
rmse_text = 'RMSE:\n' + str(np.round(_rms['rmse'], 3)) + ' +- ' + str(np.round(_rms['std'], 3)) + 'eV/atom'
ax.text(0.9, 0.1, rmse_text, transform=ax.transAxes, fontsize='large', horizontalalignment='right',
verticalalignment='bottom')
def force_plot(in_file, out_file, ax, symbol='HO', title='Plot of force'):
""" Plots the distribution of firce components per atom on the output vs the input
only plots for the given atom type(s)"""
in_atoms = ase.io.read(in_file, ':')
out_atoms = ase.io.read(out_file, ':')
# extract data for only one species
in_force, out_force = [], []
for at_in, at_out in zip(in_atoms, out_atoms):
# get the symbols
sym_all = at_in.get_chemical_symbols()
# add force for each atom
for j, sym in enumerate(sym_all):
if sym in symbol:
in_force.append(at_in.get_forces()[j])
#out_force.append(at_out.get_forces()[j]) \
out_force.append(at_out.arrays['force'][j]) # because QUIP and ASE use different names
# convert to np arrays, much easier to work with
#in_force = np.array(in_force)
#out_force = np.array(out_force)
# scatter plot of the data
ax.scatter(in_force, out_force)
# get the appropriate limits for the plot
for_limits = np.array(in_force + out_force)
flim = (for_limits.min() - 1, for_limits.max() + 1)
ax.set_xlim(flim)
ax.set_ylim(flim)
# add line of
ax.plot(flim, flim, c='k')
# set labels
ax.set_ylabel('force by GAP / (eV/Å)')
ax.set_xlabel('force by EMT / (eV/Å)')
#set title
ax.set_title(title)
# add text about RMSE
_rms = rms_dict(in_force, out_force)
rmse_text = 'RMSE:\n' + str(np.round(_rms['rmse'], 3)) + ' +- ' + str(np.round(_rms['std'], 3)) + 'eV/Å'
ax.text(0.9, 0.1, rmse_text, transform=ax.transAxes, fontsize='large', horizontalalignment='right',
verticalalignment='bottom')
[19]:
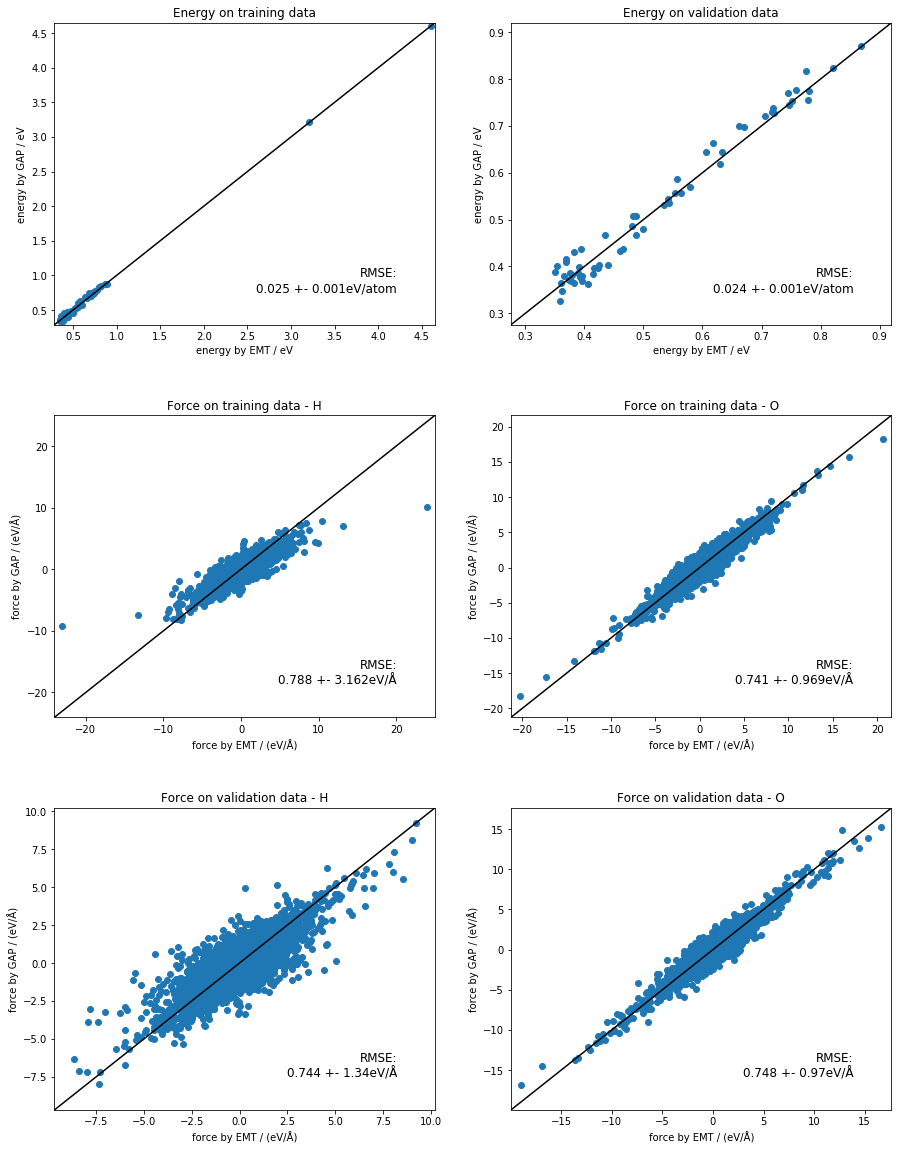
fig, ax_list = plt.subplots(nrows=3, ncols=2, gridspec_kw={'hspace': 0.3})
fig.set_size_inches(15, 20)
ax_list = ax_list.flat[:]
energy_plot('train.xyz', 'quip_train.xyz', ax_list[0], 'Energy on training data')
energy_plot('validate.xyz', 'quip_validate.xyz', ax_list[1], 'Energy on validation data')
force_plot('train.xyz', 'quip_train.xyz', ax_list[2], 'H', 'Force on training data - H')
force_plot('train.xyz', 'quip_train.xyz', ax_list[3], 'O', 'Force on training data - O')
force_plot('validate.xyz', 'quip_validate.xyz', ax_list[4], 'H', 'Force on validation data - H')
force_plot('validate.xyz', 'quip_validate.xyz', ax_list[5], 'O', 'Force on validation data - O')
# if you wanted to have the same limits on the force plots
#for ax in ax_list[2:]:
# flim = (-20, 20)
# ax.set_xlim(flim)
# ax.set_ylim(flim)
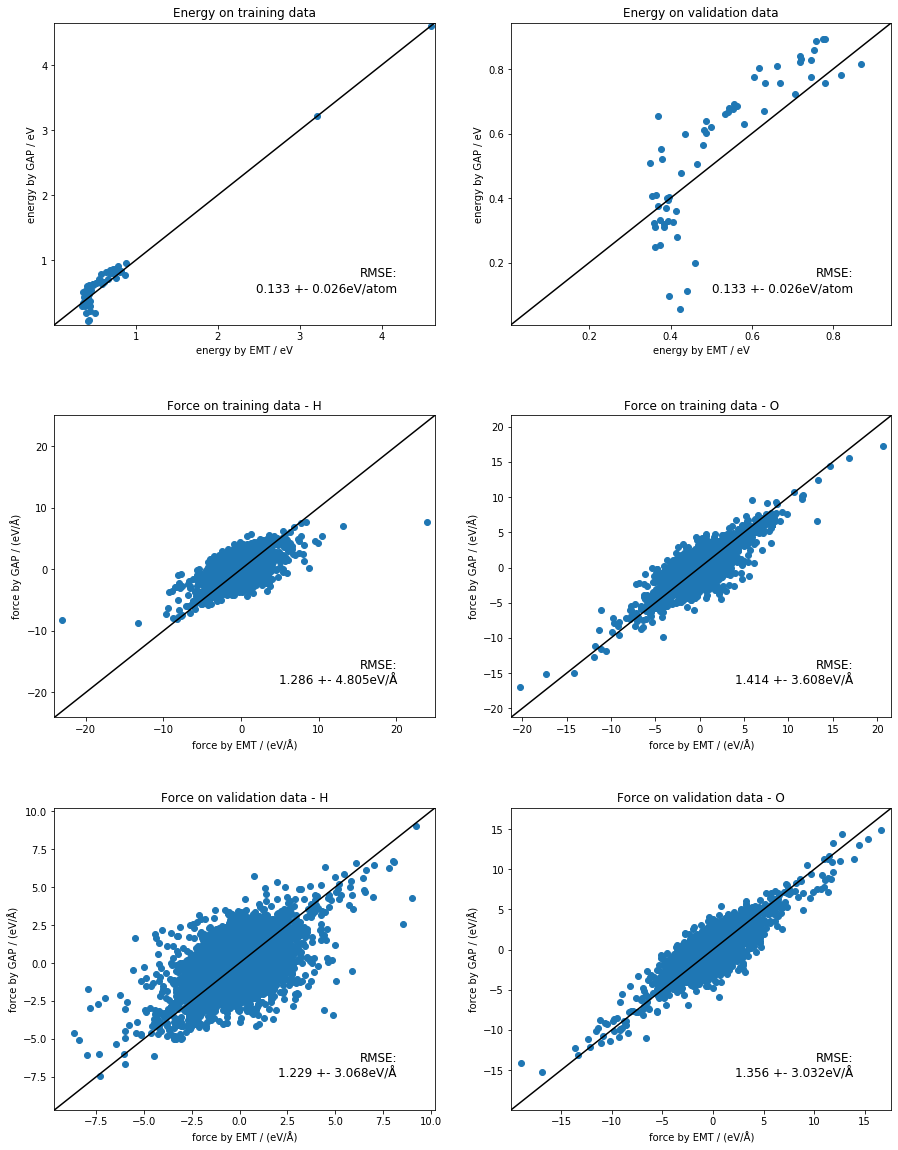
train our GAP_3b model from the command line
Let’s add three ody terms to the fit, which will hopefully improve it. We will be using the desciprtors distance_2b and angle_3b.
angle_3b - theta_fac=0.5 this takes the input data and determines the width from that; useful here, because the dimensions of the descriptor are different - n_sparse=50 higher dimensional space, more sparse points
both training and quip takes significantly more time than the last one!!!
[20]:
! gap_fit energy_parameter_name=energy force_parameter_name=forces do_copy_at_file=F sparse_separate_file=T gp_file=GAP_3b.xml at_file=train.xyz default_sigma={0.008 0.04 0 0} gap={distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform add_species=T n_sparse=10 : angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 add_species=T n_sparse=30 sparse_method=uniform}
libAtoms::Hello World: 07/10/2019 14:37:54
libAtoms::Hello World: git version git@github.com:libAtoms/QUIP.git,f538cd9fe-dirty
libAtoms::Hello World: QUIP_ARCH linux_x86_64_gfortran_openmp
libAtoms::Hello World: compiled on Oct 1 2019 at 21:54:22
libAtoms::Hello World: OpenMP parallelisation with 4 threads
WARNING: libAtoms::Hello World: environment variable OMP_STACKSIZE not set explicitly. The default value - system and compiler dependent - may be too small for some applications.
libAtoms::Hello World: Random Seed = 52674487
libAtoms::Hello World: global verbosity = 0
Calls to system_timer will do nothing by default
================================ Input parameters ==============================
at_file = train.xyz
gap = "distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform add_species=T n_sparse=10 : angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 add_species=T n_sparse=30 sparse_method=uniform"
e0 = 0.0
local_property0 = 0.0
e0_offset = 0.0
e0_method = isolated
default_sigma = "0.008 0.04 0 0"
sparse_jitter = 1.0e-10
hessian_delta = 1.0e-2
core_param_file = quip_params.xml
core_ip_args =
energy_parameter_name = energy
local_property_parameter_name = local_property
force_parameter_name = forces
virial_parameter_name = virial
hessian_parameter_name = hessian
config_type_parameter_name = config_type
sigma_parameter_name = sigma
config_type_sigma =
sigma_per_atom = T
do_copy_at_file = F
sparse_separate_file = T
sparse_use_actual_gpcov = F
gp_file = GAP_3b.xml
verbosity = NORMAL
rnd_seed = -1
do_ip_timing = F
template_file = template.xyz
sparsify_only_no_fit = F
======================================== ======================================
============== Gaussian Approximation Potentials - Database fitting ============
Initial parsing of command line arguments finished.
Found 2 GAPs.
Descriptors have been parsed
XYZ file read
Old GAP: {distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform add_species=T n_sparse=10}
New GAP: {distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=8 Z2=8}
New GAP: {distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=8 Z2=1}
New GAP: {distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=1 Z2=1}
Old GAP: { angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 add_species=T n_sparse=30 sparse_method=uniform}
New GAP: { angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=8 Z1=8 Z2=8}
New GAP: { angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=8 Z1=8 Z2=1}
New GAP: { angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=8 Z1=1 Z2=1}
New GAP: { angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=1 Z1=8 Z2=8}
New GAP: { angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=1 Z1=8 Z2=1}
New GAP: { angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=1 Z1=1 Z2=1}
Multispecies support added where requested
===================== Report on number of descriptors found ====================
---------------------------------------------------------------------
Descriptor: distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=8 Z2=8
Number of descriptors: 13192
Number of partial derivatives of descriptors: 79152
---------------------------------------------------------------------
Descriptor: distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=8 Z2=1
Number of descriptors: 63476
Number of partial derivatives of descriptors: 380856
---------------------------------------------------------------------
Descriptor: distance_2b cutoff=4.0 covariance_type=ard_se delta=0.5 theta_uniform=1.0 sparse_method=uniform n_sparse=10 Z1=1 Z2=1
Number of descriptors: 58986
Number of partial derivatives of descriptors: 353916
---------------------------------------------------------------------
Descriptor: angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=8 Z1=8 Z2=8
Number of descriptors: 49012
Number of partial derivatives of descriptors: 441108
---------------------------------------------------------------------
Descriptor: angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=8 Z1=8 Z2=1
Number of descriptors: 242862
Number of partial derivatives of descriptors: 2185758
---------------------------------------------------------------------
Descriptor: angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=8 Z1=1 Z2=1
Number of descriptors: 261574
Number of partial derivatives of descriptors: 2354166
---------------------------------------------------------------------
Descriptor: angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=1 Z1=8 Z2=8
Number of descriptors: 124922
Number of partial derivatives of descriptors: 1124298
---------------------------------------------------------------------
Descriptor: angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=1 Z1=8 Z2=1
Number of descriptors: 536486
Number of partial derivatives of descriptors: 4828374
---------------------------------------------------------------------
Descriptor: angle_3b cutoff=3.5 covariance_type=ard_se delta=0.5 theta_fac=0.5 n_sparse=30 sparse_method=uniform Z=1 Z1=1 Z2=1
Number of descriptors: 495172
Number of partial derivatives of descriptors: 4456548
======================================== ======================================
E0/atom = 0.32100000000000000E+001 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.45999999999999996E+001 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000 0.00000000000000000E+000
========== Report on number of target properties found in training XYZ: ========
Number of target energies (property name: energy) found: 63
Number of target local_properties (property name: local_property) found: 0
Number of target forces (property name: forces) found: 14829
Number of target virials (property name: virial) found: 0
Number of target Hessian eigenvalues (property name: hessian) found: 0
================================= End of report ================================
===== Report on per-configuration/per-atom sigma (error parameter) settings ====
Number of per-configuration setting of energy_sigma found: 0
Number of per-configuration setting of force_sigma found: 0
Number of per-configuration setting of virial_sigma found: 0
Number of per-configuration setting of hessian_sigma found: 0
Number of per-atom setting of force_atom_sigma found: 0
================================= End of report ================================
Cartesian coordinates transformed to descriptors
Started sparse covariance matrix calculation of coordinate 1
Finished sparse covariance matrix calculation of coordinate 1
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1_sparse done in .28807099999999952 cpu secs, .10717047499929322 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1 done in .31319400000000019 cpu secs, .11472131200025615 wall clock secs.
Started sparse covariance matrix calculation of coordinate 2
Finished sparse covariance matrix calculation of coordinate 2
TIMER: gpFull_covarianceMatrix_sparse_Coordinate2_sparse done in .59233300000000000 cpu secs, .16927113200017629 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate2 done in .59236500000000003 cpu secs, .16930502099967271 wall clock secs.
Started sparse covariance matrix calculation of coordinate 3
Finished sparse covariance matrix calculation of coordinate 3
TIMER: gpFull_covarianceMatrix_sparse_Coordinate3_sparse done in .53734700000000046 cpu secs, .15147510299993883 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate3 done in .53737699999999933 cpu secs, .15150780800013308 wall clock secs.
Started sparse covariance matrix calculation of coordinate 4
Finished sparse covariance matrix calculation of coordinate 4
TIMER: gpFull_covarianceMatrix_sparse_Coordinate4_sparse done in 1.6066120000000002 cpu secs, .42011101099978987 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate4 done in 1.6066430000000000 cpu secs, .42014362800000526 wall clock secs.
Started sparse covariance matrix calculation of coordinate 5
Finished sparse covariance matrix calculation of coordinate 5
TIMER: gpFull_covarianceMatrix_sparse_Coordinate5_sparse done in 7.3996139999999997 cpu secs, 2.0708142040002713 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate5 done in 7.3996479999999991 cpu secs, 2.0708500330001698 wall clock secs.
Started sparse covariance matrix calculation of coordinate 6
Finished sparse covariance matrix calculation of coordinate 6
TIMER: gpFull_covarianceMatrix_sparse_Coordinate6_sparse done in 7.7948529999999998 cpu secs, 2.1017793959999835 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate6 done in 7.7948830000000005 cpu secs, 2.1018120230000932 wall clock secs.
Started sparse covariance matrix calculation of coordinate 7
Finished sparse covariance matrix calculation of coordinate 7
TIMER: gpFull_covarianceMatrix_sparse_Coordinate7_sparse done in 3.7665400000000027 cpu secs, .98470028799965803 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate7 done in 3.7665709999999990 cpu secs, .98473350699987350 wall clock secs.
Started sparse covariance matrix calculation of coordinate 8
Finished sparse covariance matrix calculation of coordinate 8
TIMER: gpFull_covarianceMatrix_sparse_Coordinate8_sparse done in 15.648033999999996 cpu secs, 3.9801498979995813 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate8 done in 15.648065000000003 cpu secs, 3.9801835840007698 wall clock secs.
Started sparse covariance matrix calculation of coordinate 9
Finished sparse covariance matrix calculation of coordinate 9
TIMER: gpFull_covarianceMatrix_sparse_Coordinate9_sparse done in 14.597608999999999 cpu secs, 4.0186748889991577 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate9 done in 14.597645000000000 cpu secs, 4.0187123029991199 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_LinearAlgebra done in .58762099999999862 cpu secs, .55210713699943881 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_FunctionValues done in .19999999999242846E-004 cpu secs, .19695999981195200E-004 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse done in 52.861531999999997 cpu secs, 14.585489690999566 wall clock secs.
TIMER: GP sparsify done in 53.793665000000004 cpu secs, 15.628059424999265 wall clock secs.
libAtoms::Finalise: 07/10/2019 14:38:19
libAtoms::Finalise: Bye-Bye!
use the potential with QUIP on trani.xyz and validate.xyz
[23]:
# calculate train.xyz
! quip E=T F=T atoms_filename=train.xyz param_filename=GAP_3b.xml | grep AT | sed 's/AT//' > quip_3b_train.xyz
! quip E=T F=T atoms_filename=validate.xyz param_filename=GAP_3b.xml | grep AT | sed 's/AT//' > quip_3b_validate.xyz
look at the outputs - clear improvement
[24]:
fig, ax_list = plt.subplots(nrows=3, ncols=2, gridspec_kw={'hspace': 0.3})
fig.set_size_inches(15, 20)
ax_list = ax_list.flat[:]
energy_plot('train.xyz', 'quip_3b_train.xyz', ax_list[0], 'Energy on training data')
energy_plot('validate.xyz', 'quip_3b_validate.xyz', ax_list[1], 'Energy on validation data')
force_plot('train.xyz', 'quip_3b_train.xyz', ax_list[2], 'H', 'Force on training data - H')
force_plot('train.xyz', 'quip_3b_train.xyz', ax_list[3], 'O', 'Force on training data - O')
force_plot('validate.xyz', 'quip_3b_validate.xyz', ax_list[4], 'H', 'Force on validation data - H')
force_plot('validate.xyz', 'quip_3b_validate.xyz', ax_list[5], 'O', 'Force on validation data - O')
# if you wanted to have the same limits on the force plots
#for ax in ax_list[2:]:
# flim = (-20, 20)
# ax.set_xlim(flim)
# ax.set_ylim(flim)