The SOAP descriptor, Gaussian Approximation Potentials (GAP) and machine-learning of force fields
Adam Fekete, Matrina Stella, Henry Lambert, Alessandro De Vita, Gabor Csanyi (gc121@cam.ac.uk)
Introduction
In this tutorial we will be using a Gaussian Approximation Potentials to analyse results of TB DFT calculations of Si surface. Along the way we will learn about different descriptors (2b, 3b, soap) to describe local atomic environment in order to predict energies and forces of Si surface.
[1]:
import os
import numpy as np
import matplotlib.pylab as plt
from quippy.potential import Potential
from quippy.descriptors import Descriptor
from ase import Atoms, units
from ase.build import add_vacuum
from ase.lattice.cubic import Diamond
from ase.io import write
from ase.constraints import FixAtoms
from ase.md.velocitydistribution import MaxwellBoltzmannDistribution
from ase.md.verlet import VelocityVerlet
from ase.md.langevin import Langevin
from ase.optimize.precon import PreconLBFGS, Exp
from gap_si_surface import ViewStructure
Part I: MD, descriptors
Parameters
We can define some parameters here for the MD calculations:
[2]:
T = 1000.0 # Temperature [Kelvin]
timestep = 1.0 * units.fs
System
Defining configuration for Si slab
[3]:
def build_slab(size=(1,2,2), vacuum=10.):
# Build Si lattice.
# lattice = Diamond('Si', directions=([1, 0, 0], [0, 1, 0], [0, 0, 1]), latticeconstant=5.44, size=size)
lattice = Diamond('Si', latticeconstant=5.44, size=size)
atoms = Atoms(lattice)
# Fixing the bottom layer
bottom = atoms.positions[:,2].min()
fixed_mask = (abs(atoms.positions[:,2] - bottom) < 2.0)
atoms.set_constraint(FixAtoms(mask=fixed_mask))
# build surface by adding space to z direction
add_vacuum(atoms, vacuum)
# atoms.center(vacuum=10.0, axis=2)
return atoms
atoms = build_slab()
print('Number of atoms:', len(atoms))
Number of atoms: 32
Let’s visualise the initial configuration:
[4]:
view = ViewStructure(atoms, repetition=(2,2,1))
view
MD dynamics with TB DFTB
We use TB DFTB potential to calculate the potential energies and forces for the each timestep:
[5]:
# attach tight binding calculator
qm_pot = Potential('TB DFTB', param_filename='dftb-params.xml')
atoms.set_calculator(qm_pot)
# Thermalize atoms
MaxwellBoltzmannDistribution(atoms, 2.0 * T * units.kB)
# dynamics = VelocityVerlet(atoms, timestep)
dynamics = Langevin(atoms, timestep, T * units.kB, 0.002)
#attach observer to dynamics to track quantity of interest (temperature)
def print_status():
print('Step = {}, time = {} [fs], T = {} [K]'.format(
dynamics.nsteps,
dynamics.nsteps * dynamics.dt / units.fs,
atoms.get_kinetic_energy() / (1.5 * units.kB * len(atoms))
))
def print_energy(a=atoms): # store a reference to atoms in the definition.
"""Function to print the potential, kinetic and total energy."""
epot = a.get_potential_energy() / len(a)
ekin = a.get_kinetic_energy() / len(a)
print('Energy per atom: Epot = %.3feV Ekin = %.3feV (T=%3.0fK) '
'Etot = %.3feV' % (epot, ekin, ekin / (1.5 * units.kB), epot + ekin))
dynamics.attach(print_status, interval=10)
dynamics.attach(print_energy, interval=1)
Let’s do the dynamics:
[6]:
dynamics.run(steps=100)
# for _ in range(150):
# dynamics.run(steps=1)
Step = 0, time = 0.0 [fs], T = 2126.2241342902184 [K]
Energy per atom: Epot = -34.783eV Ekin = 0.275eV (T=2126K) Etot = -34.509eV
Energy per atom: Epot = -34.782eV Ekin = 0.263eV (T=2037K) Etot = -34.519eV
Energy per atom: Epot = -34.778eV Ekin = 0.260eV (T=2010K) Etot = -34.519eV
Energy per atom: Epot = -34.772eV Ekin = 0.253eV (T=1960K) Etot = -34.519eV
Energy per atom: Epot = -34.764eV Ekin = 0.244eV (T=1891K) Etot = -34.520eV
Energy per atom: Epot = -34.754eV Ekin = 0.234eV (T=1810K) Etot = -34.520eV
Energy per atom: Epot = -34.743eV Ekin = 0.222eV (T=1721K) Etot = -34.521eV
Energy per atom: Epot = -34.731eV Ekin = 0.211eV (T=1633K) Etot = -34.520eV
Energy per atom: Epot = -34.718eV Ekin = 0.197eV (T=1522K) Etot = -34.521eV
Energy per atom: Epot = -34.704eV Ekin = 0.183eV (T=1418K) Etot = -34.521eV
Step = 10, time = 10.0 [fs], T = 1307.506208654726 [K]
Energy per atom: Epot = -34.690eV Ekin = 0.169eV (T=1308K) Etot = -34.521eV
Energy per atom: Epot = -34.676eV Ekin = 0.155eV (T=1196K) Etot = -34.521eV
Energy per atom: Epot = -34.662eV Ekin = 0.140eV (T=1083K) Etot = -34.522eV
Energy per atom: Epot = -34.648eV Ekin = 0.127eV (T=983K) Etot = -34.521eV
Energy per atom: Epot = -34.635eV Ekin = 0.114eV (T=883K) Etot = -34.521eV
Energy per atom: Epot = -34.623eV Ekin = 0.102eV (T=789K) Etot = -34.521eV
Energy per atom: Epot = -34.612eV Ekin = 0.092eV (T=710K) Etot = -34.520eV
Energy per atom: Epot = -34.603eV Ekin = 0.082eV (T=636K) Etot = -34.521eV
Energy per atom: Epot = -34.595eV Ekin = 0.074eV (T=574K) Etot = -34.520eV
Energy per atom: Epot = -34.588eV Ekin = 0.068eV (T=522K) Etot = -34.521eV
Step = 20, time = 20.0 [fs], T = 482.4657605134864 [K]
Energy per atom: Epot = -34.583eV Ekin = 0.062eV (T=482K) Etot = -34.521eV
Energy per atom: Epot = -34.580eV Ekin = 0.059eV (T=453K) Etot = -34.521eV
Energy per atom: Epot = -34.578eV Ekin = 0.057eV (T=437K) Etot = -34.521eV
Energy per atom: Epot = -34.578eV Ekin = 0.056eV (T=434K) Etot = -34.522eV
Energy per atom: Epot = -34.579eV Ekin = 0.057eV (T=440K) Etot = -34.522eV
Energy per atom: Epot = -34.581eV Ekin = 0.059eV (T=457K) Etot = -34.522eV
Energy per atom: Epot = -34.584eV Ekin = 0.063eV (T=484K) Etot = -34.522eV
Energy per atom: Epot = -34.589eV Ekin = 0.066eV (T=512K) Etot = -34.522eV
Energy per atom: Epot = -34.594eV Ekin = 0.071eV (T=550K) Etot = -34.523eV
Energy per atom: Epot = -34.600eV Ekin = 0.077eV (T=596K) Etot = -34.523eV
Step = 30, time = 30.0 [fs], T = 641.2190478661748 [K]
Energy per atom: Epot = -34.606eV Ekin = 0.083eV (T=641K) Etot = -34.523eV
Energy per atom: Epot = -34.613eV Ekin = 0.090eV (T=696K) Etot = -34.523eV
Energy per atom: Epot = -34.620eV Ekin = 0.097eV (T=753K) Etot = -34.523eV
Energy per atom: Epot = -34.627eV Ekin = 0.104eV (T=807K) Etot = -34.523eV
Energy per atom: Epot = -34.635eV Ekin = 0.111eV (T=863K) Etot = -34.523eV
Energy per atom: Epot = -34.642eV Ekin = 0.119eV (T=919K) Etot = -34.523eV
Energy per atom: Epot = -34.649eV Ekin = 0.126eV (T=972K) Etot = -34.523eV
Energy per atom: Epot = -34.656eV Ekin = 0.132eV (T=1025K) Etot = -34.524eV
Energy per atom: Epot = -34.663eV Ekin = 0.140eV (T=1079K) Etot = -34.523eV
Energy per atom: Epot = -34.669eV Ekin = 0.145eV (T=1121K) Etot = -34.524eV
Step = 40, time = 40.0 [fs], T = 1161.738945086739 [K]
Energy per atom: Epot = -34.674eV Ekin = 0.150eV (T=1162K) Etot = -34.524eV
Energy per atom: Epot = -34.680eV Ekin = 0.155eV (T=1201K) Etot = -34.524eV
Energy per atom: Epot = -34.684eV Ekin = 0.160eV (T=1240K) Etot = -34.524eV
Energy per atom: Epot = -34.688eV Ekin = 0.164eV (T=1271K) Etot = -34.524eV
Energy per atom: Epot = -34.691eV Ekin = 0.168eV (T=1297K) Etot = -34.524eV
Energy per atom: Epot = -34.694eV Ekin = 0.172eV (T=1330K) Etot = -34.522eV
Energy per atom: Epot = -34.696eV Ekin = 0.175eV (T=1353K) Etot = -34.521eV
Energy per atom: Epot = -34.698eV Ekin = 0.177eV (T=1371K) Etot = -34.520eV
Energy per atom: Epot = -34.698eV Ekin = 0.178eV (T=1375K) Etot = -34.521eV
Energy per atom: Epot = -34.699eV Ekin = 0.178eV (T=1378K) Etot = -34.521eV
Step = 50, time = 50.00000000000001 [fs], T = 1373.781903076175 [K]
Energy per atom: Epot = -34.698eV Ekin = 0.178eV (T=1374K) Etot = -34.521eV
Energy per atom: Epot = -34.698eV Ekin = 0.176eV (T=1364K) Etot = -34.521eV
Energy per atom: Epot = -34.696eV Ekin = 0.176eV (T=1360K) Etot = -34.521eV
Energy per atom: Epot = -34.695eV Ekin = 0.175eV (T=1355K) Etot = -34.520eV
Energy per atom: Epot = -34.693eV Ekin = 0.174eV (T=1344K) Etot = -34.519eV
Energy per atom: Epot = -34.691eV Ekin = 0.171eV (T=1323K) Etot = -34.520eV
Energy per atom: Epot = -34.689eV Ekin = 0.168eV (T=1302K) Etot = -34.520eV
Energy per atom: Epot = -34.687eV Ekin = 0.165eV (T=1275K) Etot = -34.522eV
Energy per atom: Epot = -34.685eV Ekin = 0.163eV (T=1257K) Etot = -34.522eV
Energy per atom: Epot = -34.683eV Ekin = 0.161eV (T=1243K) Etot = -34.522eV
Step = 60, time = 60.0 [fs], T = 1234.8228544898413 [K]
Energy per atom: Epot = -34.682eV Ekin = 0.160eV (T=1235K) Etot = -34.522eV
Energy per atom: Epot = -34.681eV Ekin = 0.159eV (T=1229K) Etot = -34.522eV
Energy per atom: Epot = -34.680eV Ekin = 0.159eV (T=1232K) Etot = -34.521eV
Energy per atom: Epot = -34.680eV Ekin = 0.158eV (T=1219K) Etot = -34.522eV
Energy per atom: Epot = -34.680eV Ekin = 0.157eV (T=1217K) Etot = -34.522eV
Energy per atom: Epot = -34.680eV Ekin = 0.157eV (T=1217K) Etot = -34.523eV
Energy per atom: Epot = -34.681eV Ekin = 0.157eV (T=1214K) Etot = -34.524eV
Energy per atom: Epot = -34.682eV Ekin = 0.158eV (T=1219K) Etot = -34.524eV
Energy per atom: Epot = -34.683eV Ekin = 0.160eV (T=1235K) Etot = -34.523eV
Energy per atom: Epot = -34.684eV Ekin = 0.160eV (T=1241K) Etot = -34.524eV
Step = 70, time = 70.0 [fs], T = 1252.5792913035993 [K]
Energy per atom: Epot = -34.685eV Ekin = 0.162eV (T=1253K) Etot = -34.524eV
Energy per atom: Epot = -34.687eV Ekin = 0.163eV (T=1263K) Etot = -34.523eV
Energy per atom: Epot = -34.688eV Ekin = 0.165eV (T=1273K) Etot = -34.523eV
Energy per atom: Epot = -34.689eV Ekin = 0.166eV (T=1282K) Etot = -34.523eV
Energy per atom: Epot = -34.690eV Ekin = 0.168eV (T=1300K) Etot = -34.522eV
Energy per atom: Epot = -34.691eV Ekin = 0.170eV (T=1313K) Etot = -34.521eV
Energy per atom: Epot = -34.692eV Ekin = 0.171eV (T=1320K) Etot = -34.521eV
Energy per atom: Epot = -34.693eV Ekin = 0.171eV (T=1327K) Etot = -34.521eV
Energy per atom: Epot = -34.693eV Ekin = 0.172eV (T=1334K) Etot = -34.521eV
Energy per atom: Epot = -34.694eV Ekin = 0.174eV (T=1345K) Etot = -34.520eV
Step = 80, time = 80.0 [fs], T = 1354.1064927471066 [K]
Energy per atom: Epot = -34.695eV Ekin = 0.175eV (T=1354K) Etot = -34.520eV
Energy per atom: Epot = -34.696eV Ekin = 0.176eV (T=1359K) Etot = -34.520eV
Energy per atom: Epot = -34.697eV Ekin = 0.177eV (T=1373K) Etot = -34.519eV
Energy per atom: Epot = -34.698eV Ekin = 0.178eV (T=1380K) Etot = -34.520eV
Energy per atom: Epot = -34.700eV Ekin = 0.180eV (T=1390K) Etot = -34.520eV
Energy per atom: Epot = -34.701eV Ekin = 0.182eV (T=1405K) Etot = -34.520eV
Energy per atom: Epot = -34.703eV Ekin = 0.184eV (T=1420K) Etot = -34.520eV
Energy per atom: Epot = -34.705eV Ekin = 0.187eV (T=1446K) Etot = -34.519eV
Energy per atom: Epot = -34.708eV Ekin = 0.190eV (T=1466K) Etot = -34.518eV
Energy per atom: Epot = -34.711eV Ekin = 0.193eV (T=1490K) Etot = -34.518eV
Step = 90, time = 90.0 [fs], T = 1516.9294941034934 [K]
Energy per atom: Epot = -34.713eV Ekin = 0.196eV (T=1517K) Etot = -34.517eV
Energy per atom: Epot = -34.716eV Ekin = 0.199eV (T=1536K) Etot = -34.518eV
Energy per atom: Epot = -34.719eV Ekin = 0.201eV (T=1555K) Etot = -34.518eV
Energy per atom: Epot = -34.721eV Ekin = 0.203eV (T=1569K) Etot = -34.518eV
Energy per atom: Epot = -34.723eV Ekin = 0.205eV (T=1587K) Etot = -34.518eV
Energy per atom: Epot = -34.724eV Ekin = 0.206eV (T=1596K) Etot = -34.518eV
Energy per atom: Epot = -34.725eV Ekin = 0.207eV (T=1598K) Etot = -34.519eV
Energy per atom: Epot = -34.726eV Ekin = 0.207eV (T=1604K) Etot = -34.519eV
Energy per atom: Epot = -34.726eV Ekin = 0.207eV (T=1600K) Etot = -34.519eV
Energy per atom: Epot = -34.725eV Ekin = 0.207eV (T=1599K) Etot = -34.518eV
Step = 100, time = 100.00000000000001 [fs], T = 1584.1973113365673 [K]
Energy per atom: Epot = -34.723eV Ekin = 0.205eV (T=1584K) Etot = -34.518eV
[6]:
True
[8]:
view = ViewStructure(atoms)
view
Using descriptors
In this section we will intorduce some descriptors which could be useful for describing local atomic enviroments
You can find more details in the descriptor tutorial.
Pairwise
Here we use a simple pair distance between Silicon atoms, with a cutoff of 4.0 Angstrom. There are several descriptors that can do this, one is distance_2b, which takes a cutoff argument. Alternatively, the distance_Nb descriptor could also do this, with order=2. This is more general, order=3 is a triangle-like three-body descriptor of the three sides of a triangle of 3 atoms.
[7]:
desc = Descriptor("distance_Nb order=2 cutoff=4.0 n_Z=1 Zs={14}")
This descriptor is very simple: it is scalar (dimension 1), and hence only has a two (but equivalent) permutations.
[9]:
desc.n_perm
[9]:
2
[10]:
desc.cutoff()
[10]:
4.0
[11]:
desc.permutations() # array of permutation arrays
[11]:
array([[1],
[1]], dtype=int32)
Many descriptors rely on the neighbour connectivity, so we need to call calc_connect, after setting the Atoms cutoff:
[12]:
desc.cutoff()
[12]:
4.0
We can now calculate how many instances of this descriptor are found in an Atoms (or ASEAtoms) object:
[13]:
desc.count(atoms)
[13]:
195
This also works transparently for iterables (such as lists), returning a list of the counts.
We can also calculate the actual descriptor values – in this case, the list of pairwise distances:
[14]:
d = desc.calc(atoms)
# for k,v in d.items():
# print(k)
# print(type(v))
d
[14]:
{'covariance_cutoff': array([1.00000000e+00, 2.14658119e-01, 2.14658119e-01, 3.19795982e-01,
6.76831904e-01, 2.14658119e-01, 2.14658119e-01, 1.00000000e+00,
1.92216892e-01, 1.00000000e+00, 2.14658119e-01, 2.14658119e-01,
1.00000000e+00, 5.49473573e-01, 1.02669964e-01, 1.00000000e+00,
2.14658119e-01, 2.14658119e-01, 4.28565697e-03, 1.00000000e+00,
5.32303157e-01, 5.65893474e-02, 3.17080099e-02, 2.14658119e-01,
2.14658119e-01, 7.67705096e-02, 3.88763394e-01, 1.00000000e+00,
2.05886791e-01, 5.65694781e-02, 1.00000000e+00, 2.14658119e-01,
2.14658119e-01, 1.00000000e+00, 3.82420324e-01, 1.00000000e+00,
1.54120671e-01, 4.61381664e-01, 1.00000000e+00, 2.79930187e-01,
3.74608882e-02, 1.00000000e+00, 5.43735112e-01, 2.22676798e-01,
1.00000000e+00, 1.94241936e-03, 9.23567078e-02, 2.15915262e-01,
3.36721166e-01, 1.00000000e+00, 1.21132692e-01, 1.00000000e+00,
3.58304355e-07, 1.00000000e+00, 2.24123678e-01, 2.77061373e-01,
2.80646672e-01, 1.98700480e-01, 7.47843261e-03, 3.55383746e-01,
4.82663392e-01, 4.63363201e-01, 1.00000000e+00, 7.31119629e-01,
8.12121535e-01, 1.29910737e-04, 1.00000000e+00, 5.64990697e-01,
1.00000000e+00, 2.94266750e-01, 1.00000000e+00, 2.14658119e-01,
2.14658119e-01, 2.41870408e-01, 1.93580589e-01, 3.00766038e-01,
1.00000000e+00, 2.14658119e-01, 2.14658119e-01, 1.00000000e+00,
2.45323210e-01, 1.35810322e-01, 1.00000000e+00, 4.99366690e-01,
1.00000000e+00, 4.07558015e-01, 2.48598177e-01, 2.40033659e-01,
5.30349415e-02, 1.00000000e+00, 1.21403216e-03, 5.31963558e-01,
1.00000000e+00, 4.37361512e-01, 4.86924235e-01, 6.35407188e-04,
1.95029239e-01, 2.01308916e-01, 2.54279797e-01, 1.00000000e+00,
2.13940396e-02, 7.81557677e-01, 3.52627916e-01, 1.00000000e+00,
5.13536101e-01, 3.56120536e-03, 1.00000000e+00, 3.41233014e-01,
1.00000000e+00, 9.33811941e-02, 4.52377357e-01, 1.73666341e-01,
3.69737937e-01, 1.00000000e+00, 7.95747496e-01, 1.00000000e+00,
8.94305809e-01, 1.00000000e+00, 8.04260023e-01, 8.00779320e-02,
8.54949876e-04, 9.38301918e-01, 1.00000000e+00, 1.00000000e+00,
4.36305075e-01, 1.00000000e+00, 5.94272437e-01, 9.55462366e-01,
1.00000000e+00, 4.98640668e-02, 1.33190168e-01, 1.00000000e+00,
5.08484433e-02, 3.89366516e-01, 3.49320447e-01, 8.44153921e-03,
3.14779079e-02, 6.41430079e-01, 1.00000000e+00, 8.59094150e-02,
1.00000000e+00, 1.00000000e+00, 9.91037595e-01, 1.00000000e+00,
1.00000000e+00, 1.00000000e+00, 8.45588201e-04, 1.00000000e+00,
4.74170888e-01, 5.87505224e-01, 1.00000000e+00, 1.00000000e+00,
1.69943827e-01, 3.45605498e-03, 1.00000000e+00, 4.54327107e-02,
2.86809177e-01, 5.51354402e-02, 1.00000000e+00, 9.76859456e-01,
1.69917382e-01, 1.00000000e+00, 1.00000000e+00, 1.90527971e-01,
1.00000000e+00, 1.11961385e-01, 3.22921131e-01, 2.01862861e-01,
9.90932063e-01, 7.02045644e-04, 1.00000000e+00, 7.00340799e-01,
9.03175475e-01, 1.00000000e+00, 1.00000000e+00, 3.64596170e-01,
1.00000000e+00, 1.00000000e+00, 1.00000000e+00, 5.65314580e-02,
2.78732909e-03, 8.27119934e-01, 9.07878534e-02, 1.00000000e+00,
1.00000000e+00, 1.00000000e+00, 1.00000000e+00, 4.67164136e-01,
5.46809634e-01, 1.00000000e+00, 1.00000000e+00, 1.00000000e+00,
3.36143932e-01, 1.00000000e+00, 1.00000000e+00]),
'data': array([[2.3555891 ],
[3.84666089],
[3.84666089],
[3.80868127],
[3.69246785],
[3.84666089],
[3.84666089],
[2.3555891 ],
[3.85553632],
[2.3555891 ],
[3.84666089],
[3.84666089],
[2.22217899],
[3.73422626],
[3.89617545],
[2.49277209],
[3.84666089],
[3.84666089],
[3.97914695],
[2.3555891 ],
[3.73971042],
[3.92354581],
[3.94301548],
[3.84666089],
[3.84666089],
[3.91063498],
[3.7857065 ],
[2.36400224],
[3.85008681],
[3.9235595 ],
[2.3555891 ],
[3.84666089],
[3.84666089],
[2.40113644],
[3.7877806 ],
[2.53888377],
[3.8715819 ],
[3.76230485],
[2.33638577],
[3.82253553],
[3.93800043],
[2.60488918],
[3.73606087],
[3.84357323],
[2.26074713],
[3.98596662],
[3.9017103 ],
[3.8461741 ],
[3.80294437],
[2.47055579],
[3.88684692],
[2.62747264],
[3.99980946],
[2.36771171],
[3.84302037],
[3.82355411],
[3.82228164],
[3.85293408],
[3.97243879],
[3.79670004],
[3.75551952],
[3.76167232],
[2.31773498],
[3.67352306],
[3.64270374],
[3.99637188],
[2.40507651],
[3.72925412],
[2.31755331],
[3.81749164],
[2.3555891 ],
[3.84666089],
[3.84666089],
[3.83633794],
[3.85498626],
[3.81522899],
[2.3555891 ],
[3.84666089],
[3.84666089],
[2.30939588],
[3.83505773],
[3.87986306],
[2.32869864],
[3.75020159],
[2.3555891 ],
[3.77959547],
[3.83384906],
[3.83702149],
[3.92603149],
[2.36423329],
[3.98890689],
[3.73981874],
[2.27209347],
[3.76999098],
[3.75416262],
[3.99197542],
[3.85440354],
[3.85189622],
[3.83176472],
[2.39676208],
[3.95327413],
[3.65480133],
[3.79761721],
[2.22444622],
[3.7456908 ],
[3.9809933 ],
[2.49686031],
[3.80142738],
[2.19958857],
[3.90114853],
[3.76518177],
[3.86317503],
[3.79194768],
[2.25435087],
[3.64926897],
[2.25125141],
[3.60540013],
[2.36655307],
[3.64588175],
[3.90867685],
[3.99069144],
[3.57990177],
[2.49540074],
[2.33260675],
[3.77032997],
[2.57787592],
[3.71981145],
[3.56768484],
[2.45117234],
[3.92831618],
[3.8810853 ],
[2.34442646],
[3.92759974],
[3.78550961],
[3.79872015],
[3.97071307],
[3.94322485],
[3.70435838],
[2.38005406],
[3.90531209],
[3.33083172],
[2.59003818],
[3.53017959],
[2.29630022],
[2.74641565],
[2.19697139],
[3.99074256],
[2.79562658],
[3.75822532],
[3.72200204],
[2.42958642],
[2.24231016],
[3.86474567],
[3.98127633],
[2.16858341],
[3.93162786],
[3.82010597],
[3.92455341],
[3.28211822],
[3.54861007],
[3.86475688],
[2.37536444],
[2.17973943],
[3.85621962],
[2.50414849],
[3.89139667],
[3.8076162 ],
[3.85167646],
[3.53035729],
[3.99156503],
[2.23884873],
[3.68438668],
[3.60071965],
[2.22916798],
[3.21656615],
[3.79364537],
[2.65405253],
[2.70398075],
[2.36119845],
[3.9235857 ],
[3.98318696],
[3.63649434],
[3.90257603],
[2.24663903],
[3.1391255 ],
[3.44058309],
[2.17928047],
[3.76045951],
[3.73507818],
[2.88693146],
[2.27590978],
[3.39213771],
[3.80313881],
[3.14584918],
[2.44039581]]),
'has_data': array([1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1])}
Notice that the first array is called cutoff, and it supplies the value of a cutoff function, implicit in all descriptors, which takes the descriptor value to zero as the atoms approach the cutoff, i.e. in this case as the distance between the two atoms approaches the cutoff. It is more complicated for three-body and higher-body descriptors, but the final result is always a descriptor which changes smoothly with atomic positions.
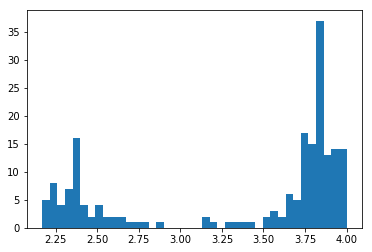
Here is a histogram of the resulting descriptor array, i.e. of the interatomic distances
[15]:
plt.hist(d['data'], bins=40)
plt.show()
Calculate size of descriptor data:
[16]:
desc.sizes(atoms)
[16]:
(195, 390)
[17]:
n_desc, n_cross = desc.sizes(atoms)
print("n_desc=%d n_cross=%d" % (n_desc, n_cross))
n_desc=195 n_cross=390
[18]:
plt.matshow(d['data'])
[18]:
<matplotlib.image.AxesImage at 0x11ce3d2e8>

SOAP descriptor
We will describe the environment using the SOAP descriptor. The SOAP descriptor of an atomic environment is based on a spherical harmonic expansion of the neighbour density, and truncating this expansion at some maximum numer of radial (n_max) and angular (l_max) indices gives rise to some parameters. We also need to give the cutoff within which we consider the neighbour environment.
Writing the descriptor vector as \(p_{ss'nn'l}\), where \(s\) and \(s'\) are indices that run over the different atomic species in the atom’s environment, \(n\) and \(n'\) are radial and \(l\) is an angular index.
[19]:
desc = Descriptor("soap cutoff=4 l_max=3 n_max=4 normalize=T atom_sigma=0.5 n_Z=1 Z={14} ")
There are now only 32 descriptors, because SOAP produces one for each atom in the structure
[20]:
desc.sizes(atoms)
[20]:
(32, 422)
But each descriptor now is a long vector, because it encodes the entire environment of the atom up to the cutoff. The length of the vector depends on l_max and n_max and also on the number of atom types.
[21]:
d = desc.calc(atoms)
d
[21]:
{'covariance_cutoff': array([1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.]),
'data': array([[8.72484873e-01, 1.34159734e-04, 1.36705926e-04, ...,
2.59809734e-08, 1.14112318e-08, 0.00000000e+00],
[7.59438258e-01, 2.64988180e-05, 2.06399928e-05, ...,
1.47470244e-08, 3.98299197e-09, 0.00000000e+00],
[8.98417344e-01, 1.38097131e-04, 1.40770407e-04, ...,
1.20256730e-08, 2.19962174e-09, 0.00000000e+00],
...,
[5.38846271e-01, 2.19806777e-05, 2.34697590e-04, ...,
1.34797058e-07, 1.20892097e-07, 0.00000000e+00],
[7.09890531e-01, 7.00599810e-05, 1.33968447e-04, ...,
4.63712579e-08, 2.52251585e-08, 0.00000000e+00],
[7.21058218e-01, 8.57141183e-05, 5.79089330e-05, ...,
8.82576223e-08, 3.07146036e-08, 0.00000000e+00]]),
'has_data': array([1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1])}
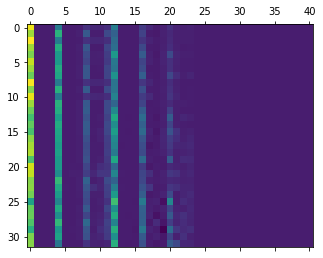
We can visualise the values of the descriptor to have an idea of how vector looks like:
[22]:
plt.matshow(d['data'])
[22]:
<matplotlib.image.AxesImage at 0x11d05a7b8>
Part II: GAP: fitting, comparing different descriptors
Database
Let’s run the MD to generate configurations for the training:
[25]:
db = []
def collect_data():
db.append(atoms.copy())
dynamics.attach(collect_data, interval=10)
dynamics.run(steps=500)
Energy per atom: Epot = -34.670eV Ekin = 0.142eV (T=1100K) Etot = -34.528eV
Energy per atom: Epot = -34.671eV Ekin = 0.143eV (T=1106K) Etot = -34.528eV
Energy per atom: Epot = -34.673eV Ekin = 0.146eV (T=1127K) Etot = -34.528eV
Energy per atom: Epot = -34.675eV Ekin = 0.147eV (T=1140K) Etot = -34.528eV
Energy per atom: Epot = -34.677eV Ekin = 0.149eV (T=1152K) Etot = -34.528eV
Energy per atom: Epot = -34.679eV Ekin = 0.151eV (T=1169K) Etot = -34.528eV
Energy per atom: Epot = -34.681eV Ekin = 0.153eV (T=1181K) Etot = -34.528eV
Energy per atom: Epot = -34.682eV Ekin = 0.154eV (T=1188K) Etot = -34.529eV
Energy per atom: Epot = -34.684eV Ekin = 0.155eV (T=1202K) Etot = -34.529eV
Step = 610, time = 610.0 [fs], T = 1213.4776009435961 [K]
Energy per atom: Epot = -34.685eV Ekin = 0.157eV (T=1213K) Etot = -34.529eV
Energy per atom: Epot = -34.687eV Ekin = 0.159eV (T=1228K) Etot = -34.528eV
Energy per atom: Epot = -34.688eV Ekin = 0.161eV (T=1242K) Etot = -34.527eV
Energy per atom: Epot = -34.689eV Ekin = 0.162eV (T=1250K) Etot = -34.527eV
Energy per atom: Epot = -34.690eV Ekin = 0.161eV (T=1248K) Etot = -34.528eV
Energy per atom: Epot = -34.690eV Ekin = 0.161eV (T=1245K) Etot = -34.529eV
Energy per atom: Epot = -34.690eV Ekin = 0.161eV (T=1249K) Etot = -34.529eV
Energy per atom: Epot = -34.690eV Ekin = 0.163eV (T=1258K) Etot = -34.527eV
Energy per atom: Epot = -34.689eV Ekin = 0.162eV (T=1254K) Etot = -34.527eV
Energy per atom: Epot = -34.689eV Ekin = 0.160eV (T=1241K) Etot = -34.528eV
Step = 620, time = 620.0 [fs], T = 1234.2040826926918 [K]
Energy per atom: Epot = -34.688eV Ekin = 0.160eV (T=1234K) Etot = -34.528eV
Energy per atom: Epot = -34.686eV Ekin = 0.159eV (T=1228K) Etot = -34.528eV
Energy per atom: Epot = -34.685eV Ekin = 0.157eV (T=1212K) Etot = -34.528eV
Energy per atom: Epot = -34.683eV Ekin = 0.156eV (T=1204K) Etot = -34.527eV
Energy per atom: Epot = -34.680eV Ekin = 0.152eV (T=1179K) Etot = -34.528eV
Energy per atom: Epot = -34.678eV Ekin = 0.151eV (T=1166K) Etot = -34.527eV
Energy per atom: Epot = -34.675eV Ekin = 0.148eV (T=1145K) Etot = -34.527eV
Energy per atom: Epot = -34.673eV Ekin = 0.145eV (T=1121K) Etot = -34.528eV
Energy per atom: Epot = -34.670eV Ekin = 0.143eV (T=1103K) Etot = -34.527eV
Energy per atom: Epot = -34.667eV Ekin = 0.140eV (T=1085K) Etot = -34.527eV
Step = 630, time = 630.0 [fs], T = 1069.8250497127774 [K]
Energy per atom: Epot = -34.665eV Ekin = 0.138eV (T=1070K) Etot = -34.527eV
Energy per atom: Epot = -34.663eV Ekin = 0.136eV (T=1054K) Etot = -34.527eV
Energy per atom: Epot = -34.661eV Ekin = 0.135eV (T=1043K) Etot = -34.527eV
Energy per atom: Epot = -34.660eV Ekin = 0.133eV (T=1025K) Etot = -34.528eV
Energy per atom: Epot = -34.659eV Ekin = 0.131eV (T=1014K) Etot = -34.528eV
Energy per atom: Epot = -34.659eV Ekin = 0.130eV (T=1009K) Etot = -34.528eV
Energy per atom: Epot = -34.659eV Ekin = 0.130eV (T=1009K) Etot = -34.529eV
Energy per atom: Epot = -34.660eV Ekin = 0.131eV (T=1015K) Etot = -34.528eV
Energy per atom: Epot = -34.661eV Ekin = 0.132eV (T=1024K) Etot = -34.528eV
Energy per atom: Epot = -34.663eV Ekin = 0.134eV (T=1040K) Etot = -34.528eV
Step = 640, time = 640.0 [fs], T = 1061.3536445382422 [K]
Energy per atom: Epot = -34.665eV Ekin = 0.137eV (T=1061K) Etot = -34.528eV
Energy per atom: Epot = -34.667eV Ekin = 0.140eV (T=1081K) Etot = -34.528eV
Energy per atom: Epot = -34.670eV Ekin = 0.143eV (T=1108K) Etot = -34.527eV
Energy per atom: Epot = -34.674eV Ekin = 0.146eV (T=1132K) Etot = -34.527eV
Energy per atom: Epot = -34.677eV Ekin = 0.150eV (T=1160K) Etot = -34.527eV
Energy per atom: Epot = -34.681eV Ekin = 0.155eV (T=1199K) Etot = -34.526eV
Energy per atom: Epot = -34.684eV Ekin = 0.158eV (T=1225K) Etot = -34.526eV
Energy per atom: Epot = -34.687eV Ekin = 0.162eV (T=1254K) Etot = -34.525eV
Energy per atom: Epot = -34.690eV Ekin = 0.164eV (T=1273K) Etot = -34.525eV
Energy per atom: Epot = -34.693eV Ekin = 0.168eV (T=1296K) Etot = -34.525eV
Step = 650, time = 650.0 [fs], T = 1316.4452722651693 [K]
Energy per atom: Epot = -34.695eV Ekin = 0.170eV (T=1316K) Etot = -34.525eV
Energy per atom: Epot = -34.697eV Ekin = 0.172eV (T=1328K) Etot = -34.525eV
Energy per atom: Epot = -34.698eV Ekin = 0.173eV (T=1339K) Etot = -34.525eV
Energy per atom: Epot = -34.699eV Ekin = 0.173eV (T=1337K) Etot = -34.526eV
Energy per atom: Epot = -34.700eV Ekin = 0.173eV (T=1342K) Etot = -34.526eV
Energy per atom: Epot = -34.700eV Ekin = 0.174eV (T=1343K) Etot = -34.526eV
Energy per atom: Epot = -34.700eV Ekin = 0.174eV (T=1347K) Etot = -34.526eV
Energy per atom: Epot = -34.699eV Ekin = 0.174eV (T=1344K) Etot = -34.525eV
Energy per atom: Epot = -34.698eV Ekin = 0.173eV (T=1339K) Etot = -34.525eV
Energy per atom: Epot = -34.697eV Ekin = 0.171eV (T=1327K) Etot = -34.525eV
Step = 660, time = 660.0 [fs], T = 1309.8201527012045 [K]
Energy per atom: Epot = -34.695eV Ekin = 0.169eV (T=1310K) Etot = -34.526eV
Energy per atom: Epot = -34.693eV Ekin = 0.166eV (T=1286K) Etot = -34.527eV
Energy per atom: Epot = -34.691eV Ekin = 0.165eV (T=1274K) Etot = -34.526eV
Energy per atom: Epot = -34.689eV Ekin = 0.162eV (T=1254K) Etot = -34.527eV
Energy per atom: Epot = -34.687eV Ekin = 0.160eV (T=1237K) Etot = -34.527eV
Energy per atom: Epot = -34.684eV Ekin = 0.159eV (T=1227K) Etot = -34.526eV
Energy per atom: Epot = -34.682eV Ekin = 0.156eV (T=1206K) Etot = -34.526eV
Energy per atom: Epot = -34.680eV Ekin = 0.154eV (T=1190K) Etot = -34.526eV
Energy per atom: Epot = -34.678eV Ekin = 0.152eV (T=1175K) Etot = -34.526eV
Energy per atom: Epot = -34.676eV Ekin = 0.151eV (T=1170K) Etot = -34.525eV
Step = 670, time = 670.0 [fs], T = 1161.851603014357 [K]
Energy per atom: Epot = -34.675eV Ekin = 0.150eV (T=1162K) Etot = -34.525eV
Energy per atom: Epot = -34.673eV Ekin = 0.148eV (T=1149K) Etot = -34.525eV
Energy per atom: Epot = -34.672eV Ekin = 0.147eV (T=1137K) Etot = -34.525eV
Energy per atom: Epot = -34.671eV Ekin = 0.147eV (T=1137K) Etot = -34.524eV
Energy per atom: Epot = -34.670eV Ekin = 0.145eV (T=1123K) Etot = -34.525eV
Energy per atom: Epot = -34.669eV Ekin = 0.144eV (T=1116K) Etot = -34.525eV
Energy per atom: Epot = -34.669eV Ekin = 0.143eV (T=1106K) Etot = -34.526eV
Energy per atom: Epot = -34.669eV Ekin = 0.143eV (T=1104K) Etot = -34.526eV
Energy per atom: Epot = -34.669eV Ekin = 0.143eV (T=1106K) Etot = -34.526eV
Energy per atom: Epot = -34.669eV Ekin = 0.143eV (T=1109K) Etot = -34.525eV
Step = 680, time = 680.0 [fs], T = 1116.049981841541 [K]
Energy per atom: Epot = -34.669eV Ekin = 0.144eV (T=1116K) Etot = -34.525eV
Energy per atom: Epot = -34.669eV Ekin = 0.145eV (T=1120K) Etot = -34.525eV
Energy per atom: Epot = -34.670eV Ekin = 0.146eV (T=1129K) Etot = -34.524eV
Energy per atom: Epot = -34.670eV Ekin = 0.147eV (T=1138K) Etot = -34.523eV
Energy per atom: Epot = -34.671eV Ekin = 0.148eV (T=1144K) Etot = -34.523eV
Energy per atom: Epot = -34.671eV Ekin = 0.147eV (T=1137K) Etot = -34.524eV
Energy per atom: Epot = -34.671eV Ekin = 0.146eV (T=1133K) Etot = -34.525eV
Energy per atom: Epot = -34.672eV Ekin = 0.147eV (T=1134K) Etot = -34.525eV
Energy per atom: Epot = -34.671eV Ekin = 0.146eV (T=1133K) Etot = -34.525eV
Energy per atom: Epot = -34.671eV Ekin = 0.147eV (T=1136K) Etot = -34.524eV
Step = 690, time = 689.9999999999999 [fs], T = 1138.1637640732908 [K]
Energy per atom: Epot = -34.671eV Ekin = 0.147eV (T=1138K) Etot = -34.524eV
Energy per atom: Epot = -34.670eV Ekin = 0.146eV (T=1129K) Etot = -34.524eV
Energy per atom: Epot = -34.670eV Ekin = 0.145eV (T=1120K) Etot = -34.525eV
Energy per atom: Epot = -34.669eV Ekin = 0.145eV (T=1120K) Etot = -34.524eV
Energy per atom: Epot = -34.668eV Ekin = 0.144eV (T=1112K) Etot = -34.524eV
Energy per atom: Epot = -34.667eV Ekin = 0.142eV (T=1102K) Etot = -34.524eV
Energy per atom: Epot = -34.666eV Ekin = 0.140eV (T=1084K) Etot = -34.526eV
Energy per atom: Epot = -34.665eV Ekin = 0.140eV (T=1084K) Etot = -34.525eV
Energy per atom: Epot = -34.664eV Ekin = 0.139eV (T=1072K) Etot = -34.525eV
Energy per atom: Epot = -34.663eV Ekin = 0.137eV (T=1062K) Etot = -34.526eV
Step = 700, time = 700.0000000000001 [fs], T = 1054.1133918643613 [K]
Energy per atom: Epot = -34.663eV Ekin = 0.136eV (T=1054K) Etot = -34.526eV
Energy per atom: Epot = -34.662eV Ekin = 0.137eV (T=1057K) Etot = -34.525eV
Energy per atom: Epot = -34.662eV Ekin = 0.135eV (T=1044K) Etot = -34.527eV
Energy per atom: Epot = -34.661eV Ekin = 0.135eV (T=1044K) Etot = -34.526eV
Energy per atom: Epot = -34.661eV Ekin = 0.135eV (T=1047K) Etot = -34.526eV
Energy per atom: Epot = -34.661eV Ekin = 0.136eV (T=1051K) Etot = -34.526eV
Energy per atom: Epot = -34.662eV Ekin = 0.136eV (T=1056K) Etot = -34.525eV
Energy per atom: Epot = -34.662eV Ekin = 0.137eV (T=1062K) Etot = -34.525eV
Energy per atom: Epot = -34.663eV Ekin = 0.138eV (T=1065K) Etot = -34.526eV
Energy per atom: Epot = -34.664eV Ekin = 0.139eV (T=1075K) Etot = -34.525eV
Step = 710, time = 710.0 [fs], T = 1087.5729168770392 [K]
Energy per atom: Epot = -34.666eV Ekin = 0.141eV (T=1088K) Etot = -34.525eV
Energy per atom: Epot = -34.667eV Ekin = 0.143eV (T=1106K) Etot = -34.524eV
Energy per atom: Epot = -34.669eV Ekin = 0.144eV (T=1116K) Etot = -34.525eV
Energy per atom: Epot = -34.670eV Ekin = 0.146eV (T=1130K) Etot = -34.524eV
Energy per atom: Epot = -34.672eV Ekin = 0.148eV (T=1148K) Etot = -34.524eV
Energy per atom: Epot = -34.674eV Ekin = 0.151eV (T=1167K) Etot = -34.523eV
Energy per atom: Epot = -34.676eV Ekin = 0.153eV (T=1181K) Etot = -34.524eV
Energy per atom: Epot = -34.678eV Ekin = 0.155eV (T=1203K) Etot = -34.523eV
Energy per atom: Epot = -34.680eV Ekin = 0.158eV (T=1225K) Etot = -34.522eV
Energy per atom: Epot = -34.682eV Ekin = 0.161eV (T=1244K) Etot = -34.521eV
Step = 720, time = 720.0 [fs], T = 1260.4486838568973 [K]
Energy per atom: Epot = -34.684eV Ekin = 0.163eV (T=1260K) Etot = -34.521eV
Energy per atom: Epot = -34.686eV Ekin = 0.164eV (T=1271K) Etot = -34.522eV
Energy per atom: Epot = -34.688eV Ekin = 0.167eV (T=1289K) Etot = -34.521eV
Energy per atom: Epot = -34.689eV Ekin = 0.167eV (T=1292K) Etot = -34.522eV
Energy per atom: Epot = -34.690eV Ekin = 0.168eV (T=1302K) Etot = -34.522eV
Energy per atom: Epot = -34.691eV Ekin = 0.170eV (T=1318K) Etot = -34.520eV
Energy per atom: Epot = -34.691eV Ekin = 0.171eV (T=1321K) Etot = -34.521eV
Energy per atom: Epot = -34.691eV Ekin = 0.171eV (T=1319K) Etot = -34.521eV
Energy per atom: Epot = -34.691eV Ekin = 0.170eV (T=1313K) Etot = -34.521eV
Energy per atom: Epot = -34.691eV Ekin = 0.170eV (T=1313K) Etot = -34.521eV
Step = 730, time = 730.0 [fs], T = 1306.1395297984811 [K]
Energy per atom: Epot = -34.690eV Ekin = 0.169eV (T=1306K) Etot = -34.521eV
Energy per atom: Epot = -34.688eV Ekin = 0.168eV (T=1297K) Etot = -34.521eV
Energy per atom: Epot = -34.687eV Ekin = 0.166eV (T=1285K) Etot = -34.521eV
Energy per atom: Epot = -34.685eV Ekin = 0.164eV (T=1270K) Etot = -34.521eV
Energy per atom: Epot = -34.683eV Ekin = 0.162eV (T=1255K) Etot = -34.521eV
Energy per atom: Epot = -34.680eV Ekin = 0.160eV (T=1241K) Etot = -34.520eV
Energy per atom: Epot = -34.678eV Ekin = 0.158eV (T=1220K) Etot = -34.520eV
Energy per atom: Epot = -34.675eV Ekin = 0.154eV (T=1194K) Etot = -34.521eV
Energy per atom: Epot = -34.673eV Ekin = 0.151eV (T=1169K) Etot = -34.522eV
Energy per atom: Epot = -34.671eV Ekin = 0.149eV (T=1155K) Etot = -34.521eV
Step = 740, time = 740.0 [fs], T = 1130.0228639362892 [K]
Energy per atom: Epot = -34.668eV Ekin = 0.146eV (T=1130K) Etot = -34.522eV
Energy per atom: Epot = -34.666eV Ekin = 0.144eV (T=1113K) Etot = -34.522eV
Energy per atom: Epot = -34.665eV Ekin = 0.142eV (T=1102K) Etot = -34.522eV
Energy per atom: Epot = -34.663eV Ekin = 0.141eV (T=1089K) Etot = -34.523eV
Energy per atom: Epot = -34.662eV Ekin = 0.140eV (T=1083K) Etot = -34.523eV
Energy per atom: Epot = -34.662eV Ekin = 0.140eV (T=1084K) Etot = -34.522eV
Energy per atom: Epot = -34.662eV Ekin = 0.140eV (T=1079K) Etot = -34.522eV
Energy per atom: Epot = -34.662eV Ekin = 0.139eV (T=1073K) Etot = -34.523eV
Energy per atom: Epot = -34.663eV Ekin = 0.138eV (T=1069K) Etot = -34.524eV
Energy per atom: Epot = -34.664eV Ekin = 0.139eV (T=1076K) Etot = -34.525eV
Step = 750, time = 750.0 [fs], T = 1083.9614905678948 [K]
Energy per atom: Epot = -34.665eV Ekin = 0.140eV (T=1084K) Etot = -34.525eV
Energy per atom: Epot = -34.666eV Ekin = 0.141eV (T=1092K) Etot = -34.525eV
Energy per atom: Epot = -34.668eV Ekin = 0.143eV (T=1105K) Etot = -34.525eV
Energy per atom: Epot = -34.670eV Ekin = 0.145eV (T=1118K) Etot = -34.525eV
Energy per atom: Epot = -34.672eV Ekin = 0.147eV (T=1140K) Etot = -34.525eV
Energy per atom: Epot = -34.674eV Ekin = 0.149eV (T=1149K) Etot = -34.525eV
Energy per atom: Epot = -34.676eV Ekin = 0.150eV (T=1158K) Etot = -34.526eV
Energy per atom: Epot = -34.677eV Ekin = 0.150eV (T=1162K) Etot = -34.527eV
Energy per atom: Epot = -34.679eV Ekin = 0.151eV (T=1172K) Etot = -34.527eV
Energy per atom: Epot = -34.680eV Ekin = 0.152eV (T=1174K) Etot = -34.528eV
Step = 760, time = 760.0 [fs], T = 1182.2959304995202 [K]
Energy per atom: Epot = -34.681eV Ekin = 0.153eV (T=1182K) Etot = -34.528eV
Energy per atom: Epot = -34.681eV Ekin = 0.154eV (T=1188K) Etot = -34.528eV
Energy per atom: Epot = -34.681eV Ekin = 0.152eV (T=1180K) Etot = -34.529eV
Energy per atom: Epot = -34.681eV Ekin = 0.152eV (T=1175K) Etot = -34.529eV
Energy per atom: Epot = -34.681eV Ekin = 0.151eV (T=1168K) Etot = -34.530eV
Energy per atom: Epot = -34.680eV Ekin = 0.150eV (T=1162K) Etot = -34.530eV
Energy per atom: Epot = -34.679eV Ekin = 0.148eV (T=1147K) Etot = -34.530eV
Energy per atom: Epot = -34.677eV Ekin = 0.147eV (T=1134K) Etot = -34.531eV
Energy per atom: Epot = -34.676eV Ekin = 0.144eV (T=1117K) Etot = -34.531eV
Energy per atom: Epot = -34.674eV Ekin = 0.142eV (T=1100K) Etot = -34.532eV
Step = 770, time = 770.0 [fs], T = 1089.9711537191097 [K]
Energy per atom: Epot = -34.672eV Ekin = 0.141eV (T=1090K) Etot = -34.531eV
Energy per atom: Epot = -34.670eV Ekin = 0.140eV (T=1080K) Etot = -34.531eV
Energy per atom: Epot = -34.669eV Ekin = 0.138eV (T=1069K) Etot = -34.531eV
Energy per atom: Epot = -34.667eV Ekin = 0.136eV (T=1054K) Etot = -34.531eV
Energy per atom: Epot = -34.666eV Ekin = 0.134eV (T=1035K) Etot = -34.532eV
Energy per atom: Epot = -34.665eV Ekin = 0.134eV (T=1038K) Etot = -34.531eV
Energy per atom: Epot = -34.664eV Ekin = 0.134eV (T=1033K) Etot = -34.530eV
Energy per atom: Epot = -34.664eV Ekin = 0.133eV (T=1030K) Etot = -34.531eV
Energy per atom: Epot = -34.664eV Ekin = 0.134eV (T=1040K) Etot = -34.529eV
Energy per atom: Epot = -34.665eV Ekin = 0.135eV (T=1046K) Etot = -34.529eV
Step = 780, time = 780.0 [fs], T = 1058.2368819251544 [K]
Energy per atom: Epot = -34.666eV Ekin = 0.137eV (T=1058K) Etot = -34.529eV
Energy per atom: Epot = -34.667eV Ekin = 0.139eV (T=1072K) Etot = -34.529eV
Energy per atom: Epot = -34.669eV Ekin = 0.141eV (T=1091K) Etot = -34.528eV
Energy per atom: Epot = -34.671eV Ekin = 0.143eV (T=1109K) Etot = -34.528eV
Energy per atom: Epot = -34.674eV Ekin = 0.145eV (T=1125K) Etot = -34.528eV
Energy per atom: Epot = -34.676eV Ekin = 0.148eV (T=1147K) Etot = -34.528eV
Energy per atom: Epot = -34.678eV Ekin = 0.151eV (T=1169K) Etot = -34.527eV
Energy per atom: Epot = -34.681eV Ekin = 0.152eV (T=1178K) Etot = -34.529eV
Energy per atom: Epot = -34.683eV Ekin = 0.153eV (T=1186K) Etot = -34.530eV
Energy per atom: Epot = -34.685eV Ekin = 0.156eV (T=1205K) Etot = -34.530eV
Step = 790, time = 789.9999999999999 [fs], T = 1221.4561259539792 [K]
Energy per atom: Epot = -34.687eV Ekin = 0.158eV (T=1221K) Etot = -34.529eV
Energy per atom: Epot = -34.688eV Ekin = 0.159eV (T=1227K) Etot = -34.530eV
Energy per atom: Epot = -34.689eV Ekin = 0.160eV (T=1236K) Etot = -34.530eV
Energy per atom: Epot = -34.690eV Ekin = 0.161eV (T=1245K) Etot = -34.529eV
Energy per atom: Epot = -34.690eV Ekin = 0.162eV (T=1250K) Etot = -34.529eV
Energy per atom: Epot = -34.690eV Ekin = 0.161eV (T=1249K) Etot = -34.529eV
Energy per atom: Epot = -34.690eV Ekin = 0.162eV (T=1251K) Etot = -34.528eV
Energy per atom: Epot = -34.689eV Ekin = 0.161eV (T=1248K) Etot = -34.528eV
Energy per atom: Epot = -34.688eV Ekin = 0.161eV (T=1244K) Etot = -34.527eV
Energy per atom: Epot = -34.687eV Ekin = 0.160eV (T=1240K) Etot = -34.526eV
Step = 800, time = 800.0000000000001 [fs], T = 1224.3944703568234 [K]
Energy per atom: Epot = -34.685eV Ekin = 0.158eV (T=1224K) Etot = -34.527eV
Energy per atom: Epot = -34.683eV Ekin = 0.156eV (T=1204K) Etot = -34.528eV
Energy per atom: Epot = -34.681eV Ekin = 0.153eV (T=1185K) Etot = -34.528eV
Energy per atom: Epot = -34.679eV Ekin = 0.152eV (T=1174K) Etot = -34.527eV
Energy per atom: Epot = -34.676eV Ekin = 0.149eV (T=1154K) Etot = -34.527eV
Energy per atom: Epot = -34.674eV Ekin = 0.147eV (T=1136K) Etot = -34.527eV
Energy per atom: Epot = -34.671eV Ekin = 0.143eV (T=1108K) Etot = -34.528eV
Energy per atom: Epot = -34.669eV Ekin = 0.141eV (T=1089K) Etot = -34.528eV
Energy per atom: Epot = -34.666eV Ekin = 0.139eV (T=1074K) Etot = -34.528eV
Energy per atom: Epot = -34.664eV Ekin = 0.136eV (T=1056K) Etot = -34.528eV
Step = 810, time = 810.0 [fs], T = 1048.562618524431 [K]
Energy per atom: Epot = -34.662eV Ekin = 0.136eV (T=1049K) Etot = -34.527eV
Energy per atom: Epot = -34.660eV Ekin = 0.133eV (T=1029K) Etot = -34.527eV
Energy per atom: Epot = -34.659eV Ekin = 0.131eV (T=1014K) Etot = -34.528eV
Energy per atom: Epot = -34.658eV Ekin = 0.130eV (T=1003K) Etot = -34.528eV
Energy per atom: Epot = -34.658eV Ekin = 0.129eV (T=999K) Etot = -34.529eV
Energy per atom: Epot = -34.658eV Ekin = 0.129eV (T=1001K) Etot = -34.528eV
Energy per atom: Epot = -34.658eV Ekin = 0.130eV (T=1009K) Etot = -34.528eV
Energy per atom: Epot = -34.659eV Ekin = 0.131eV (T=1013K) Etot = -34.528eV
Energy per atom: Epot = -34.660eV Ekin = 0.132eV (T=1024K) Etot = -34.527eV
Energy per atom: Epot = -34.661eV Ekin = 0.134eV (T=1038K) Etot = -34.527eV
Step = 820, time = 820.0 [fs], T = 1047.2420327306986 [K]
Energy per atom: Epot = -34.663eV Ekin = 0.135eV (T=1047K) Etot = -34.527eV
Energy per atom: Epot = -34.665eV Ekin = 0.137eV (T=1062K) Etot = -34.527eV
Energy per atom: Epot = -34.667eV Ekin = 0.140eV (T=1081K) Etot = -34.527eV
Energy per atom: Epot = -34.669eV Ekin = 0.142eV (T=1101K) Etot = -34.527eV
Energy per atom: Epot = -34.672eV Ekin = 0.144eV (T=1118K) Etot = -34.527eV
Energy per atom: Epot = -34.674eV Ekin = 0.147eV (T=1136K) Etot = -34.527eV
Energy per atom: Epot = -34.677eV Ekin = 0.150eV (T=1162K) Etot = -34.527eV
Energy per atom: Epot = -34.680eV Ekin = 0.153eV (T=1187K) Etot = -34.526eV
Energy per atom: Epot = -34.683eV Ekin = 0.155eV (T=1199K) Etot = -34.528eV
Energy per atom: Epot = -34.686eV Ekin = 0.159eV (T=1233K) Etot = -34.527eV
Step = 830, time = 830.0 [fs], T = 1261.8494846560873 [K]
Energy per atom: Epot = -34.689eV Ekin = 0.163eV (T=1262K) Etot = -34.526eV
Energy per atom: Epot = -34.693eV Ekin = 0.166eV (T=1286K) Etot = -34.526eV
Energy per atom: Epot = -34.696eV Ekin = 0.171eV (T=1319K) Etot = -34.525eV
Energy per atom: Epot = -34.699eV Ekin = 0.174eV (T=1345K) Etot = -34.525eV
Energy per atom: Epot = -34.702eV Ekin = 0.176eV (T=1362K) Etot = -34.526eV
Energy per atom: Epot = -34.705eV Ekin = 0.179eV (T=1386K) Etot = -34.526eV
Energy per atom: Epot = -34.707eV Ekin = 0.182eV (T=1409K) Etot = -34.525eV
Energy per atom: Epot = -34.710eV Ekin = 0.185eV (T=1428K) Etot = -34.525eV
Energy per atom: Epot = -34.712eV Ekin = 0.187eV (T=1446K) Etot = -34.525eV
Energy per atom: Epot = -34.713eV Ekin = 0.189eV (T=1464K) Etot = -34.524eV
Step = 840, time = 840.0 [fs], T = 1473.1796543543905 [K]
Energy per atom: Epot = -34.714eV Ekin = 0.190eV (T=1473K) Etot = -34.524eV
Energy per atom: Epot = -34.714eV Ekin = 0.191eV (T=1477K) Etot = -34.523eV
Energy per atom: Epot = -34.714eV Ekin = 0.192eV (T=1484K) Etot = -34.522eV
Energy per atom: Epot = -34.713eV Ekin = 0.191eV (T=1476K) Etot = -34.522eV
Energy per atom: Epot = -34.712eV Ekin = 0.191eV (T=1475K) Etot = -34.521eV
Energy per atom: Epot = -34.710eV Ekin = 0.188eV (T=1454K) Etot = -34.522eV
Energy per atom: Epot = -34.707eV Ekin = 0.185eV (T=1429K) Etot = -34.522eV
Energy per atom: Epot = -34.704eV Ekin = 0.181eV (T=1401K) Etot = -34.523eV
Energy per atom: Epot = -34.701eV Ekin = 0.177eV (T=1371K) Etot = -34.524eV
Energy per atom: Epot = -34.698eV Ekin = 0.174eV (T=1345K) Etot = -34.524eV
Step = 850, time = 850.0 [fs], T = 1319.0981610208323 [K]
Energy per atom: Epot = -34.695eV Ekin = 0.171eV (T=1319K) Etot = -34.524eV
Energy per atom: Epot = -34.691eV Ekin = 0.167eV (T=1295K) Etot = -34.524eV
Energy per atom: Epot = -34.688eV Ekin = 0.164eV (T=1271K) Etot = -34.524eV
Energy per atom: Epot = -34.686eV Ekin = 0.162eV (T=1254K) Etot = -34.523eV
Energy per atom: Epot = -34.683eV Ekin = 0.160eV (T=1236K) Etot = -34.524eV
Energy per atom: Epot = -34.682eV Ekin = 0.158eV (T=1222K) Etot = -34.524eV
Energy per atom: Epot = -34.681eV Ekin = 0.157eV (T=1211K) Etot = -34.524eV
Energy per atom: Epot = -34.681eV Ekin = 0.156eV (T=1207K) Etot = -34.525eV
Energy per atom: Epot = -34.681eV Ekin = 0.156eV (T=1208K) Etot = -34.525eV
Energy per atom: Epot = -34.682eV Ekin = 0.158eV (T=1219K) Etot = -34.524eV
Step = 860, time = 860.0 [fs], T = 1230.7107298901024 [K]
Energy per atom: Epot = -34.683eV Ekin = 0.159eV (T=1231K) Etot = -34.524eV
Energy per atom: Epot = -34.685eV Ekin = 0.160eV (T=1237K) Etot = -34.525eV
Energy per atom: Epot = -34.688eV Ekin = 0.162eV (T=1252K) Etot = -34.526eV
Energy per atom: Epot = -34.691eV Ekin = 0.165eV (T=1279K) Etot = -34.525eV
Energy per atom: Epot = -34.694eV Ekin = 0.168eV (T=1303K) Etot = -34.525eV
Energy per atom: Epot = -34.697eV Ekin = 0.171eV (T=1325K) Etot = -34.526eV
Energy per atom: Epot = -34.700eV Ekin = 0.175eV (T=1352K) Etot = -34.526eV
Energy per atom: Epot = -34.704eV Ekin = 0.178eV (T=1375K) Etot = -34.526eV
Energy per atom: Epot = -34.707eV Ekin = 0.181eV (T=1400K) Etot = -34.526eV
Energy per atom: Epot = -34.710eV Ekin = 0.184eV (T=1426K) Etot = -34.526eV
Step = 870, time = 870.0 [fs], T = 1445.5688460227605 [K]
Energy per atom: Epot = -34.713eV Ekin = 0.187eV (T=1446K) Etot = -34.526eV
Energy per atom: Epot = -34.715eV Ekin = 0.189eV (T=1459K) Etot = -34.527eV
Energy per atom: Epot = -34.718eV Ekin = 0.191eV (T=1480K) Etot = -34.526eV
Energy per atom: Epot = -34.720eV Ekin = 0.193eV (T=1493K) Etot = -34.527eV
Energy per atom: Epot = -34.721eV Ekin = 0.195eV (T=1506K) Etot = -34.526eV
Energy per atom: Epot = -34.722eV Ekin = 0.196eV (T=1516K) Etot = -34.526eV
Energy per atom: Epot = -34.723eV Ekin = 0.197eV (T=1524K) Etot = -34.526eV
Energy per atom: Epot = -34.724eV Ekin = 0.199eV (T=1538K) Etot = -34.525eV
Energy per atom: Epot = -34.724eV Ekin = 0.201eV (T=1553K) Etot = -34.523eV
Energy per atom: Epot = -34.723eV Ekin = 0.201eV (T=1554K) Etot = -34.522eV
Step = 880, time = 879.9999999999999 [fs], T = 1550.4222940578104 [K]
Energy per atom: Epot = -34.722eV Ekin = 0.200eV (T=1550K) Etot = -34.522eV
Energy per atom: Epot = -34.721eV Ekin = 0.198eV (T=1533K) Etot = -34.523eV
Energy per atom: Epot = -34.719eV Ekin = 0.197eV (T=1522K) Etot = -34.523eV
Energy per atom: Epot = -34.717eV Ekin = 0.195eV (T=1507K) Etot = -34.522eV
Energy per atom: Epot = -34.715eV Ekin = 0.193eV (T=1495K) Etot = -34.522eV
Energy per atom: Epot = -34.712eV Ekin = 0.191eV (T=1476K) Etot = -34.521eV
Energy per atom: Epot = -34.709eV Ekin = 0.188eV (T=1455K) Etot = -34.521eV
Energy per atom: Epot = -34.706eV Ekin = 0.184eV (T=1423K) Etot = -34.522eV
Energy per atom: Epot = -34.703eV Ekin = 0.182eV (T=1405K) Etot = -34.521eV
Energy per atom: Epot = -34.699eV Ekin = 0.178eV (T=1376K) Etot = -34.521eV
Step = 890, time = 890.0000000000001 [fs], T = 1349.3007898319372 [K]
Energy per atom: Epot = -34.695eV Ekin = 0.174eV (T=1349K) Etot = -34.521eV
Energy per atom: Epot = -34.692eV Ekin = 0.171eV (T=1321K) Etot = -34.521eV
Energy per atom: Epot = -34.688eV Ekin = 0.166eV (T=1283K) Etot = -34.522eV
Energy per atom: Epot = -34.685eV Ekin = 0.162eV (T=1252K) Etot = -34.523eV
Energy per atom: Epot = -34.681eV Ekin = 0.158eV (T=1224K) Etot = -34.523eV
Energy per atom: Epot = -34.678eV Ekin = 0.154eV (T=1195K) Etot = -34.524eV
Energy per atom: Epot = -34.675eV Ekin = 0.152eV (T=1176K) Etot = -34.523eV
Energy per atom: Epot = -34.673eV Ekin = 0.149eV (T=1155K) Etot = -34.523eV
Energy per atom: Epot = -34.670eV Ekin = 0.147eV (T=1134K) Etot = -34.524eV
Energy per atom: Epot = -34.668eV Ekin = 0.145eV (T=1125K) Etot = -34.523eV
Step = 900, time = 900.0 [fs], T = 1112.7854874238203 [K]
Energy per atom: Epot = -34.666eV Ekin = 0.144eV (T=1113K) Etot = -34.522eV
Energy per atom: Epot = -34.665eV Ekin = 0.143eV (T=1108K) Etot = -34.522eV
Energy per atom: Epot = -34.664eV Ekin = 0.143eV (T=1105K) Etot = -34.521eV
Energy per atom: Epot = -34.663eV Ekin = 0.142eV (T=1098K) Etot = -34.521eV
Energy per atom: Epot = -34.663eV Ekin = 0.141eV (T=1090K) Etot = -34.522eV
Energy per atom: Epot = -34.664eV Ekin = 0.141eV (T=1092K) Etot = -34.523eV
Energy per atom: Epot = -34.664eV Ekin = 0.142eV (T=1100K) Etot = -34.522eV
Energy per atom: Epot = -34.666eV Ekin = 0.143eV (T=1110K) Etot = -34.522eV
Energy per atom: Epot = -34.667eV Ekin = 0.145eV (T=1122K) Etot = -34.522eV
Energy per atom: Epot = -34.669eV Ekin = 0.147eV (T=1134K) Etot = -34.523eV
Step = 910, time = 910.0 [fs], T = 1154.638311116254 [K]
Energy per atom: Epot = -34.672eV Ekin = 0.149eV (T=1155K) Etot = -34.522eV
Energy per atom: Epot = -34.674eV Ekin = 0.152eV (T=1174K) Etot = -34.522eV
Energy per atom: Epot = -34.677eV Ekin = 0.154eV (T=1194K) Etot = -34.523eV
Energy per atom: Epot = -34.680eV Ekin = 0.157eV (T=1217K) Etot = -34.523eV
Energy per atom: Epot = -34.683eV Ekin = 0.161eV (T=1243K) Etot = -34.522eV
Energy per atom: Epot = -34.686eV Ekin = 0.163eV (T=1263K) Etot = -34.523eV
Energy per atom: Epot = -34.689eV Ekin = 0.167eV (T=1290K) Etot = -34.522eV
Energy per atom: Epot = -34.692eV Ekin = 0.169eV (T=1310K) Etot = -34.523eV
Energy per atom: Epot = -34.695eV Ekin = 0.172eV (T=1332K) Etot = -34.523eV
Energy per atom: Epot = -34.698eV Ekin = 0.175eV (T=1354K) Etot = -34.523eV
Step = 920, time = 920.0 [fs], T = 1375.175238346479 [K]
Energy per atom: Epot = -34.700eV Ekin = 0.178eV (T=1375K) Etot = -34.522eV
Energy per atom: Epot = -34.702eV Ekin = 0.179eV (T=1388K) Etot = -34.523eV
Energy per atom: Epot = -34.704eV Ekin = 0.182eV (T=1405K) Etot = -34.523eV
Energy per atom: Epot = -34.706eV Ekin = 0.183eV (T=1419K) Etot = -34.523eV
Energy per atom: Epot = -34.708eV Ekin = 0.186eV (T=1438K) Etot = -34.522eV
Energy per atom: Epot = -34.709eV Ekin = 0.187eV (T=1447K) Etot = -34.522eV
Energy per atom: Epot = -34.710eV Ekin = 0.188eV (T=1452K) Etot = -34.522eV
Energy per atom: Epot = -34.710eV Ekin = 0.188eV (T=1456K) Etot = -34.522eV
Energy per atom: Epot = -34.711eV Ekin = 0.189eV (T=1460K) Etot = -34.522eV
Energy per atom: Epot = -34.710eV Ekin = 0.189eV (T=1460K) Etot = -34.522eV
Step = 930, time = 930.0 [fs], T = 1458.7143296739027 [K]
Energy per atom: Epot = -34.710eV Ekin = 0.189eV (T=1459K) Etot = -34.521eV
Energy per atom: Epot = -34.709eV Ekin = 0.189eV (T=1460K) Etot = -34.520eV
Energy per atom: Epot = -34.708eV Ekin = 0.187eV (T=1444K) Etot = -34.521eV
Energy per atom: Epot = -34.707eV Ekin = 0.186eV (T=1441K) Etot = -34.520eV
Energy per atom: Epot = -34.705eV Ekin = 0.186eV (T=1437K) Etot = -34.519eV
Energy per atom: Epot = -34.703eV Ekin = 0.184eV (T=1424K) Etot = -34.519eV
Energy per atom: Epot = -34.701eV Ekin = 0.182eV (T=1405K) Etot = -34.519eV
Energy per atom: Epot = -34.698eV Ekin = 0.179eV (T=1381K) Etot = -34.520eV
Energy per atom: Epot = -34.696eV Ekin = 0.175eV (T=1351K) Etot = -34.521eV
Energy per atom: Epot = -34.693eV Ekin = 0.173eV (T=1337K) Etot = -34.520eV
Step = 940, time = 940.0 [fs], T = 1318.911914675727 [K]
Energy per atom: Epot = -34.690eV Ekin = 0.170eV (T=1319K) Etot = -34.519eV
Energy per atom: Epot = -34.686eV Ekin = 0.167eV (T=1290K) Etot = -34.520eV
Energy per atom: Epot = -34.683eV Ekin = 0.163eV (T=1262K) Etot = -34.520eV
Energy per atom: Epot = -34.680eV Ekin = 0.159eV (T=1233K) Etot = -34.520eV
Energy per atom: Epot = -34.677eV Ekin = 0.157eV (T=1212K) Etot = -34.520eV
Energy per atom: Epot = -34.674eV Ekin = 0.153eV (T=1187K) Etot = -34.520eV
Energy per atom: Epot = -34.671eV Ekin = 0.150eV (T=1164K) Etot = -34.520eV
Energy per atom: Epot = -34.668eV Ekin = 0.148eV (T=1142K) Etot = -34.521eV
Energy per atom: Epot = -34.666eV Ekin = 0.145eV (T=1124K) Etot = -34.521eV
Energy per atom: Epot = -34.664eV Ekin = 0.143eV (T=1107K) Etot = -34.521eV
Step = 950, time = 950.0 [fs], T = 1099.0873560001837 [K]
Energy per atom: Epot = -34.662eV Ekin = 0.142eV (T=1099K) Etot = -34.520eV
Energy per atom: Epot = -34.661eV Ekin = 0.141eV (T=1088K) Etot = -34.521eV
Energy per atom: Epot = -34.661eV Ekin = 0.140eV (T=1080K) Etot = -34.521eV
Energy per atom: Epot = -34.660eV Ekin = 0.140eV (T=1082K) Etot = -34.520eV
Energy per atom: Epot = -34.660eV Ekin = 0.141eV (T=1087K) Etot = -34.519eV
Energy per atom: Epot = -34.660eV Ekin = 0.141eV (T=1090K) Etot = -34.519eV
Energy per atom: Epot = -34.661eV Ekin = 0.141eV (T=1089K) Etot = -34.520eV
Energy per atom: Epot = -34.661eV Ekin = 0.141eV (T=1090K) Etot = -34.520eV
Energy per atom: Epot = -34.662eV Ekin = 0.142eV (T=1102K) Etot = -34.520eV
Energy per atom: Epot = -34.663eV Ekin = 0.144eV (T=1113K) Etot = -34.519eV
Step = 960, time = 960.0 [fs], T = 1121.7438534640573 [K]
Energy per atom: Epot = -34.664eV Ekin = 0.145eV (T=1122K) Etot = -34.519eV
Energy per atom: Epot = -34.665eV Ekin = 0.147eV (T=1136K) Etot = -34.518eV
Energy per atom: Epot = -34.666eV Ekin = 0.147eV (T=1136K) Etot = -34.519eV
Energy per atom: Epot = -34.667eV Ekin = 0.147eV (T=1135K) Etot = -34.520eV
Energy per atom: Epot = -34.668eV Ekin = 0.148eV (T=1144K) Etot = -34.520eV
Energy per atom: Epot = -34.668eV Ekin = 0.147eV (T=1140K) Etot = -34.521eV
Energy per atom: Epot = -34.669eV Ekin = 0.148eV (T=1145K) Etot = -34.521eV
Energy per atom: Epot = -34.669eV Ekin = 0.148eV (T=1144K) Etot = -34.521eV
Energy per atom: Epot = -34.669eV Ekin = 0.148eV (T=1145K) Etot = -34.521eV
Energy per atom: Epot = -34.669eV Ekin = 0.148eV (T=1144K) Etot = -34.521eV
Step = 970, time = 969.9999999999999 [fs], T = 1150.8046259890853 [K]
Energy per atom: Epot = -34.669eV Ekin = 0.149eV (T=1151K) Etot = -34.520eV
Energy per atom: Epot = -34.669eV Ekin = 0.147eV (T=1135K) Etot = -34.522eV
Energy per atom: Epot = -34.668eV Ekin = 0.146eV (T=1127K) Etot = -34.523eV
Energy per atom: Epot = -34.668eV Ekin = 0.145eV (T=1122K) Etot = -34.523eV
Energy per atom: Epot = -34.667eV Ekin = 0.144eV (T=1115K) Etot = -34.523eV
Energy per atom: Epot = -34.667eV Ekin = 0.144eV (T=1115K) Etot = -34.523eV
Energy per atom: Epot = -34.667eV Ekin = 0.144eV (T=1118K) Etot = -34.522eV
Energy per atom: Epot = -34.666eV Ekin = 0.143eV (T=1108K) Etot = -34.523eV
Energy per atom: Epot = -34.666eV Ekin = 0.143eV (T=1107K) Etot = -34.523eV
Energy per atom: Epot = -34.666eV Ekin = 0.144eV (T=1110K) Etot = -34.522eV
Step = 980, time = 980.0000000000001 [fs], T = 1116.9613964890314 [K]
Energy per atom: Epot = -34.666eV Ekin = 0.144eV (T=1117K) Etot = -34.522eV
Energy per atom: Epot = -34.667eV Ekin = 0.145eV (T=1124K) Etot = -34.521eV
Energy per atom: Epot = -34.667eV Ekin = 0.146eV (T=1132K) Etot = -34.521eV
Energy per atom: Epot = -34.668eV Ekin = 0.148eV (T=1143K) Etot = -34.521eV
Energy per atom: Epot = -34.670eV Ekin = 0.150eV (T=1162K) Etot = -34.520eV
Energy per atom: Epot = -34.671eV Ekin = 0.152eV (T=1176K) Etot = -34.519eV
Energy per atom: Epot = -34.673eV Ekin = 0.154eV (T=1194K) Etot = -34.519eV
Energy per atom: Epot = -34.675eV Ekin = 0.156eV (T=1209K) Etot = -34.519eV
Energy per atom: Epot = -34.677eV Ekin = 0.159eV (T=1228K) Etot = -34.519eV
Energy per atom: Epot = -34.680eV Ekin = 0.162eV (T=1253K) Etot = -34.518eV
Step = 990, time = 990.0 [fs], T = 1261.7042483844475 [K]
Energy per atom: Epot = -34.682eV Ekin = 0.163eV (T=1262K) Etot = -34.519eV
Energy per atom: Epot = -34.684eV Ekin = 0.165eV (T=1279K) Etot = -34.519eV
Energy per atom: Epot = -34.686eV Ekin = 0.168eV (T=1296K) Etot = -34.519eV
Energy per atom: Epot = -34.688eV Ekin = 0.170eV (T=1316K) Etot = -34.518eV
Energy per atom: Epot = -34.690eV Ekin = 0.172eV (T=1332K) Etot = -34.518eV
Energy per atom: Epot = -34.691eV Ekin = 0.173eV (T=1338K) Etot = -34.518eV
Energy per atom: Epot = -34.692eV Ekin = 0.173eV (T=1342K) Etot = -34.518eV
Energy per atom: Epot = -34.692eV Ekin = 0.174eV (T=1343K) Etot = -34.518eV
Energy per atom: Epot = -34.691eV Ekin = 0.173eV (T=1338K) Etot = -34.518eV
Energy per atom: Epot = -34.690eV Ekin = 0.172eV (T=1330K) Etot = -34.518eV
Step = 1000, time = 1000.0 [fs], T = 1312.3702881693948 [K]
Energy per atom: Epot = -34.689eV Ekin = 0.170eV (T=1312K) Etot = -34.519eV
Energy per atom: Epot = -34.687eV Ekin = 0.168eV (T=1298K) Etot = -34.519eV
Energy per atom: Epot = -34.684eV Ekin = 0.165eV (T=1280K) Etot = -34.519eV
Energy per atom: Epot = -34.681eV Ekin = 0.163eV (T=1263K) Etot = -34.518eV
Energy per atom: Epot = -34.678eV Ekin = 0.160eV (T=1237K) Etot = -34.518eV
Energy per atom: Epot = -34.675eV Ekin = 0.157eV (T=1214K) Etot = -34.518eV
Energy per atom: Epot = -34.671eV Ekin = 0.154eV (T=1190K) Etot = -34.518eV
Energy per atom: Epot = -34.668eV Ekin = 0.151eV (T=1166K) Etot = -34.518eV
Energy per atom: Epot = -34.666eV Ekin = 0.147eV (T=1139K) Etot = -34.518eV
Energy per atom: Epot = -34.663eV Ekin = 0.145eV (T=1119K) Etot = -34.519eV
Step = 1010, time = 1010.0 [fs], T = 1107.3682349275302 [K]
Energy per atom: Epot = -34.661eV Ekin = 0.143eV (T=1107K) Etot = -34.518eV
Energy per atom: Epot = -34.660eV Ekin = 0.142eV (T=1097K) Etot = -34.519eV
Energy per atom: Epot = -34.660eV Ekin = 0.142eV (T=1099K) Etot = -34.518eV
Energy per atom: Epot = -34.660eV Ekin = 0.143eV (T=1103K) Etot = -34.518eV
Energy per atom: Epot = -34.662eV Ekin = 0.143eV (T=1108K) Etot = -34.518eV
Energy per atom: Epot = -34.664eV Ekin = 0.145eV (T=1118K) Etot = -34.519eV
Energy per atom: Epot = -34.666eV Ekin = 0.147eV (T=1134K) Etot = -34.520eV
Energy per atom: Epot = -34.669eV Ekin = 0.150eV (T=1162K) Etot = -34.519eV
Energy per atom: Epot = -34.673eV Ekin = 0.154eV (T=1193K) Etot = -34.519eV
Energy per atom: Epot = -34.677eV Ekin = 0.158eV (T=1225K) Etot = -34.519eV
Step = 1020, time = 1020.0 [fs], T = 1254.456856444442 [K]
Energy per atom: Epot = -34.682eV Ekin = 0.162eV (T=1254K) Etot = -34.520eV
Energy per atom: Epot = -34.687eV Ekin = 0.166eV (T=1287K) Etot = -34.520eV
Energy per atom: Epot = -34.692eV Ekin = 0.170eV (T=1314K) Etot = -34.522eV
Energy per atom: Epot = -34.696eV Ekin = 0.175eV (T=1357K) Etot = -34.521eV
Energy per atom: Epot = -34.701eV Ekin = 0.180eV (T=1389K) Etot = -34.521eV
Energy per atom: Epot = -34.705eV Ekin = 0.184eV (T=1425K) Etot = -34.521eV
Energy per atom: Epot = -34.709eV Ekin = 0.189eV (T=1461K) Etot = -34.521eV
Energy per atom: Epot = -34.713eV Ekin = 0.191eV (T=1476K) Etot = -34.522eV
Energy per atom: Epot = -34.716eV Ekin = 0.193eV (T=1496K) Etot = -34.523eV
Energy per atom: Epot = -34.719eV Ekin = 0.196eV (T=1515K) Etot = -34.523eV
Step = 1030, time = 1030.0 [fs], T = 1533.4115403699773 [K]
Energy per atom: Epot = -34.721eV Ekin = 0.198eV (T=1533K) Etot = -34.523eV
Energy per atom: Epot = -34.722eV Ekin = 0.199eV (T=1543K) Etot = -34.523eV
Energy per atom: Epot = -34.723eV Ekin = 0.200eV (T=1548K) Etot = -34.523eV
Energy per atom: Epot = -34.724eV Ekin = 0.201eV (T=1557K) Etot = -34.522eV
Energy per atom: Epot = -34.723eV Ekin = 0.202eV (T=1559K) Etot = -34.522eV
Energy per atom: Epot = -34.723eV Ekin = 0.201eV (T=1559K) Etot = -34.521eV
Energy per atom: Epot = -34.721eV Ekin = 0.200eV (T=1544K) Etot = -34.522eV
Energy per atom: Epot = -34.720eV Ekin = 0.199eV (T=1540K) Etot = -34.521eV
Energy per atom: Epot = -34.718eV Ekin = 0.196eV (T=1519K) Etot = -34.521eV
Energy per atom: Epot = -34.715eV Ekin = 0.194eV (T=1499K) Etot = -34.522eV
Step = 1040, time = 1040.0 [fs], T = 1475.3818337384587 [K]
Energy per atom: Epot = -34.713eV Ekin = 0.191eV (T=1475K) Etot = -34.522eV
Energy per atom: Epot = -34.710eV Ekin = 0.187eV (T=1450K) Etot = -34.523eV
Energy per atom: Epot = -34.708eV Ekin = 0.185eV (T=1434K) Etot = -34.522eV
Energy per atom: Epot = -34.705eV Ekin = 0.183eV (T=1416K) Etot = -34.522eV
Energy per atom: Epot = -34.703eV Ekin = 0.180eV (T=1391K) Etot = -34.523eV
Energy per atom: Epot = -34.700eV Ekin = 0.177eV (T=1369K) Etot = -34.523eV
Energy per atom: Epot = -34.698eV Ekin = 0.176eV (T=1361K) Etot = -34.522eV
Energy per atom: Epot = -34.696eV Ekin = 0.173eV (T=1339K) Etot = -34.523eV
Energy per atom: Epot = -34.694eV Ekin = 0.171eV (T=1321K) Etot = -34.523eV
Energy per atom: Epot = -34.692eV Ekin = 0.169eV (T=1306K) Etot = -34.523eV
Step = 1050, time = 1050.0 [fs], T = 1289.440068746159 [K]
Energy per atom: Epot = -34.691eV Ekin = 0.167eV (T=1289K) Etot = -34.524eV
Energy per atom: Epot = -34.689eV Ekin = 0.164eV (T=1271K) Etot = -34.525eV
Energy per atom: Epot = -34.687eV Ekin = 0.162eV (T=1252K) Etot = -34.526eV
Energy per atom: Epot = -34.686eV Ekin = 0.160eV (T=1236K) Etot = -34.526eV
Energy per atom: Epot = -34.684eV Ekin = 0.158eV (T=1223K) Etot = -34.526eV
Energy per atom: Epot = -34.682eV Ekin = 0.156eV (T=1203K) Etot = -34.527eV
Energy per atom: Epot = -34.680eV Ekin = 0.153eV (T=1182K) Etot = -34.527eV
Energy per atom: Epot = -34.678eV Ekin = 0.151eV (T=1166K) Etot = -34.528eV
Energy per atom: Epot = -34.676eV Ekin = 0.148eV (T=1147K) Etot = -34.528eV
Energy per atom: Epot = -34.674eV Ekin = 0.146eV (T=1133K) Etot = -34.527eV
Step = 1060, time = 1060.0 [fs], T = 1106.3208287365883 [K]
Energy per atom: Epot = -34.671eV Ekin = 0.143eV (T=1106K) Etot = -34.528eV
Energy per atom: Epot = -34.669eV Ekin = 0.141eV (T=1091K) Etot = -34.528eV
Energy per atom: Epot = -34.666eV Ekin = 0.138eV (T=1069K) Etot = -34.528eV
Energy per atom: Epot = -34.664eV Ekin = 0.135eV (T=1045K) Etot = -34.529eV
Energy per atom: Epot = -34.662eV Ekin = 0.132eV (T=1025K) Etot = -34.529eV
Energy per atom: Epot = -34.660eV Ekin = 0.130eV (T=1010K) Etot = -34.529eV
Energy per atom: Epot = -34.658eV Ekin = 0.130eV (T=1002K) Etot = -34.528eV
Energy per atom: Epot = -34.656eV Ekin = 0.127eV (T=985K) Etot = -34.529eV
Energy per atom: Epot = -34.656eV Ekin = 0.127eV (T=985K) Etot = -34.528eV
Energy per atom: Epot = -34.655eV Ekin = 0.126eV (T=976K) Etot = -34.529eV
Step = 1070, time = 1070.0 [fs], T = 980.9725165651028 [K]
Energy per atom: Epot = -34.656eV Ekin = 0.127eV (T=981K) Etot = -34.529eV
Energy per atom: Epot = -34.657eV Ekin = 0.127eV (T=981K) Etot = -34.530eV
Energy per atom: Epot = -34.658eV Ekin = 0.129eV (T=994K) Etot = -34.530eV
Energy per atom: Epot = -34.660eV Ekin = 0.130eV (T=1005K) Etot = -34.530eV
Energy per atom: Epot = -34.663eV Ekin = 0.132eV (T=1023K) Etot = -34.531eV
Energy per atom: Epot = -34.666eV Ekin = 0.136eV (T=1048K) Etot = -34.530eV
Energy per atom: Epot = -34.670eV Ekin = 0.140eV (T=1081K) Etot = -34.530eV
Energy per atom: Epot = -34.674eV Ekin = 0.143eV (T=1110K) Etot = -34.530eV
Energy per atom: Epot = -34.678eV Ekin = 0.147eV (T=1139K) Etot = -34.531eV
Energy per atom: Epot = -34.683eV Ekin = 0.152eV (T=1176K) Etot = -34.531eV
Step = 1080, time = 1080.0 [fs], T = 1210.131324601977 [K]
Energy per atom: Epot = -34.687eV Ekin = 0.156eV (T=1210K) Etot = -34.531eV
Energy per atom: Epot = -34.692eV Ekin = 0.160eV (T=1241K) Etot = -34.531eV
Energy per atom: Epot = -34.696eV Ekin = 0.165eV (T=1273K) Etot = -34.531eV
Energy per atom: Epot = -34.700eV Ekin = 0.169eV (T=1311K) Etot = -34.531eV
Energy per atom: Epot = -34.704eV Ekin = 0.173eV (T=1342K) Etot = -34.531eV
Energy per atom: Epot = -34.707eV Ekin = 0.177eV (T=1369K) Etot = -34.530eV
Energy per atom: Epot = -34.710eV Ekin = 0.180eV (T=1393K) Etot = -34.530eV
Energy per atom: Epot = -34.713eV Ekin = 0.182eV (T=1411K) Etot = -34.530eV
Energy per atom: Epot = -34.715eV Ekin = 0.185eV (T=1428K) Etot = -34.530eV
Energy per atom: Epot = -34.716eV Ekin = 0.186eV (T=1437K) Etot = -34.531eV
Step = 1090, time = 1090.0 [fs], T = 1439.9055594814956 [K]
Energy per atom: Epot = -34.717eV Ekin = 0.186eV (T=1440K) Etot = -34.531eV
Energy per atom: Epot = -34.717eV Ekin = 0.185eV (T=1433K) Etot = -34.532eV
Energy per atom: Epot = -34.717eV Ekin = 0.185eV (T=1430K) Etot = -34.532eV
Energy per atom: Epot = -34.717eV Ekin = 0.183eV (T=1416K) Etot = -34.534eV
Energy per atom: Epot = -34.715eV Ekin = 0.182eV (T=1410K) Etot = -34.533eV
Energy per atom: Epot = -34.714eV Ekin = 0.181eV (T=1398K) Etot = -34.533eV
Energy per atom: Epot = -34.712eV Ekin = 0.178eV (T=1378K) Etot = -34.534eV
Energy per atom: Epot = -34.710eV Ekin = 0.176eV (T=1361K) Etot = -34.534eV
Energy per atom: Epot = -34.708eV Ekin = 0.173eV (T=1338K) Etot = -34.535eV
Energy per atom: Epot = -34.705eV Ekin = 0.170eV (T=1318K) Etot = -34.535eV
Step = 1100, time = 1100.0 [fs], T = 1294.479828341841 [K]
Energy per atom: Epot = -34.703eV Ekin = 0.167eV (T=1294K) Etot = -34.535eV
[25]:
True
[26]:
# The size of the database:
len(db)
[26]:
100
Save the results into a file for using it as an input for the fitting:
[27]:
write('/tmp/atoms_db.xyz', db)
Fitting the 2-body potential
We need to calculate the atomic energy and use it as an offset during the fitting:
[28]:
isolated_atom = Atoms("Si", positions=[[0,0,0]])
isolated_atom.set_calculator(qm_pot)
E0 = isolated_atom.get_potential_energy()
E0
[28]:
-29.716948405885105
We are going to use teach_sparse tool to build the GAP potential. For a detailed description of the available options you can run the following command.
[34]:
!gap_fit --help
libAtoms::Hello World: 07/10/2019 17:16:56
libAtoms::Hello World: git version git@github.com:/libAtoms/QUIP,d204dca49-dirty
libAtoms::Hello World: QUIP_ARCH darwin_x86_64_gfortran_openmp
libAtoms::Hello World: compiled on Aug 7 2019 at 14:13:45
libAtoms::Hello World: OpenMP parallelisation with 4 threads
WARNING: libAtoms::Hello World: environment variable OMP_STACKSIZE not set explicitly. The default value - system and compiler dependent - may be too small for some applications.
libAtoms::Hello World: Random Seed = 62216297
libAtoms::Hello World: global verbosity = 0
Calls to system_timer will do nothing by default
at_file type=STRING scalar current_value=//MANDATORY//
XYZ file with fitting configurations
gap type=STRING scalar current_value=//MANDATORY//
Initialisation string for GAPs
e0 type=STRING scalar current_value=0.0
Atomic energy value to be subtracted from energies before fitting (and added
back on after prediction). Specifiy a single number (used for all species) or
by species: {Ti:-150.0:O:-320}. energy = core + GAP +
e0
local_property0 type=STRING scalar current_value=0.0
Local property value to be subtracted from the local property before fitting
(and added back on after prediction). Specifiy a single number (used for all
species) or by species: {H:20.0:Cl:35.0}.
e0_offset type=REAL scalar current_value=0.0
Offset of baseline. If zero, the offset is the average atomic energy of the inpu-
t data or the e0 specified manually.
e0_method type=STRING scalar current_value=isolated
Method to determine e0, if not explicitly specified. Possible options: isolated
(default, each atom present in the XYZ needs to have an isolated representative,
with a valid energy), average (e0 is the average of all total energies across
the XYZ)
default_sigma type=REAL dim=4 current_value=//MANDATORY//
error in [energies forces virials hessians]
sparse_jitter type=REAL scalar current_value=1.0e-10
intrisic error of atomic/bond energy, used to regularise the sparse covariance
matrix
hessian_delta type=REAL scalar current_value=1.0e-2
Delta to use in numerical differentiation when obtaining second derivative for
the Hessian covariance
core_param_file type=STRING scalar current_value=quip_params.xml
QUIP XML file for a potential to subtract from data (and added back after predic-
tion)
core_ip_args type=STRING scalar current_value=
QUIP init string for a potential to subtract from data (and added back after
prediction)
energy_parameter_name type=STRING scalar current_value=energy
Name of energy property in the at_file that describes the data
local_property_parameter_name type=STRING scalar current_value=local_property
Name of local_property in the at_file that describes the data
force_parameter_name type=STRING scalar current_value=force
Name of force property in the at_file that describes the data
virial_parameter_name type=STRING scalar current_value=virial
Name of virial property in the at_file that describes the data
hessian_parameter_name type=STRING scalar current_value=hessian
Name of hessian property in the at_file that describes the data
config_type_parameter_name type=STRING scalar current_value=config_type
Identifier of property determining the type of input data in the at_file
sigma_parameter_name type=STRING scalar current_value=sigma
sigma parameters (error hyper) for a given configuration in the database. Overri-
des the command line sigmas. In the XYZ, it must be prepended by energy_, force_-
, virial_ or hessian_
config_type_sigma type=STRING scalar current_value=
What sigma values to choose for each type of data. Format: {type:energy:force:vi-
rial:hessian}
sigma_per_atom type=LOGICAL scalar current_value=T
Interpretation of the energy and virial sigmas specified in >>default_sigma<<
and >>config_type_sigma<<. If >>T<<, they are interpreted as per-atom errors,
and the variance will be scaled according to the number of atoms in the configur-
ation. If >>F<< they are treated as absolute errors and no scaling is performed.
NOTE: sigmas specified on a per-configuration basis (see >>sigma_parameter_name<-
<) are always absolute.
do_copy_at_file type=LOGICAL scalar current_value=T
Do copy the at_file into the GAP XML file (should be set to False for NetCDF
input).
sparse_separate_file type=LOGICAL scalar current_value=T
Save sparse coordinates data in separate file
sparse_use_actual_gpcov type=LOGICAL scalar current_value=F
Use actual GP covariance for sparsification methods
gp_file type=STRING scalar current_value=gp_new.xml
output XML file
verbosity type=STRING scalar current_value=NORMAL
Verbosity control. Options: NORMAL, VERBOSE, NERD, ANALYSIS.
rnd_seed type=INTEGER scalar current_value=-1
Random seed.
do_ip_timing type=LOGICAL scalar current_value=F
To enable or not timing of the interatomic potential.
template_file type=STRING scalar current_value=template.xyz
Template XYZ file for initialising object
sparsify_only_no_fit type=LOGICAL scalar current_value=F
If true, sparsification is done, but no fitting. print the sparse index by addin-
g print_sparse_index=file.dat to the descriptor string.
missing mandatory parameter at_file
missing mandatory parameter gap
missing mandatory parameter default_sigma
gap_fit
SYSTEM ABORT: Exit: Mandatory argument(s) missing...
Now we have to do actual fitting by running the following command:
Relevant parameters: - covariance_type: Type of covariance function to use. Available: ARD_SE, DOT_PRODUCT, BOND_REAL_SP- ACE, PP (piecewise polynomial) - theta_uniform: Set the width of Gaussians for the ARD_SE and PP kernel, same in each dimension - n_sparse: Number of sparse points to use in the sparsification of the Gaussian process - delta: Set the standard deviation of the Gaussian process. Typically this would be set to the standard deviation (i.e. root mean square) of the function that is approximated with the Gaussian process. - default_sigma: error in [energies, forces, virials, hessians]
[35]:
# !gap_fit at_file=dummy default_sigma={0 0 0 0} gap={distance_Nb delta=0.0 covariance_type=ARD_SE --help}
[36]:
!gap_fit at_file=/tmp/atoms_db.xyz \
gap={distance_Nb order=2 \
cutoff=5.0 \
covariance_type=ARD_SE \
theta_uniform=1.0 \
n_sparse=15 \
delta=1.0} \
e0=-29.716948405885105 \
default_sigma={0.01 0.5 0.0 0.0} \
do_copy_at_file=F sparse_separate_file=F \
gp_file=/tmp/gap_2b.xml
libAtoms::Hello World: 07/10/2019 17:16:59
libAtoms::Hello World: git version git@github.com:/libAtoms/QUIP,d204dca49-dirty
libAtoms::Hello World: QUIP_ARCH darwin_x86_64_gfortran_openmp
libAtoms::Hello World: compiled on Aug 7 2019 at 14:13:45
libAtoms::Hello World: OpenMP parallelisation with 4 threads
WARNING: libAtoms::Hello World: environment variable OMP_STACKSIZE not set explicitly. The default value - system and compiler dependent - may be too small for some applications.
libAtoms::Hello World: Random Seed = 62219567
libAtoms::Hello World: global verbosity = 0
Calls to system_timer will do nothing by default
================================ Input parameters ==============================
at_file = /tmp/atoms_db.xyz
gap = "distance_Nb order=2 cutoff=5.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=15 delta=1.0"
e0 = -29.716948405885105
local_property0 = 0.0
e0_offset = 0.0
e0_method = isolated
default_sigma = "0.01 0.5 0.0 0.0"
sparse_jitter = 1.0e-10
hessian_delta = 1.0e-2
core_param_file = quip_params.xml
core_ip_args =
energy_parameter_name = energy
local_property_parameter_name = local_property
force_parameter_name = force
virial_parameter_name = virial
hessian_parameter_name = hessian
config_type_parameter_name = config_type
sigma_parameter_name = sigma
config_type_sigma =
sigma_per_atom = T
do_copy_at_file = F
sparse_separate_file = F
sparse_use_actual_gpcov = F
gp_file = /tmp/gap_2b.xml
verbosity = NORMAL
rnd_seed = -1
do_ip_timing = F
template_file = template.xyz
sparsify_only_no_fit = F
======================================== ======================================
============== Gaussian Approximation Potentials - Database fitting ============
Initial parsing of command line arguments finished.
Found 1 GAPs.
Descriptors have been parsed
XYZ file read
Unchanged GAP: {distance_Nb order=2 cutoff=5.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=15 delta=1.0}
Multispecies support added where requested
===================== Report on number of descriptors found ====================
---------------------------------------------------------------------
Descriptor: distance_Nb order=2 cutoff=5.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=15 delta=1.0
Number of descriptors: 35312
Number of partial derivatives of descriptors: 0
======================================== ======================================
========== Report on number of target properties found in training XYZ: ========
Number of target energies (property name: energy) found: 100
Number of target local_properties (property name: local_property) found: 0
Number of target forces (property name: force) found: 0
Number of target virials (property name: virial) found: 0
Number of target Hessian eigenvalues (property name: hessian) found: 0
================================= End of report ================================
===== Report on per-configuration/per-atom sigma (error parameter) settings ====
Number of per-configuration setting of energy_sigma found: 0
Number of per-configuration setting of force_sigma found: 0
Number of per-configuration setting of virial_sigma found: 0
Number of per-configuration setting of hessian_sigma found: 0
Number of per-atom setting of force_atom_sigma found: 0
================================= End of report ================================
Cartesian coordinates transformed to descriptors
Started sparse covariance matrix calculation of coordinate 1
Finished sparse covariance matrix calculation of coordinate 1
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1_sparse done in 1.9402960000000000 cpu secs, .60122609138488770 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1 done in 1.9403350000000001 cpu secs, .60126590728759766 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_LinearAlgebra done in .12049999999996786E-002 cpu secs, .34060478210449219E-002 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_FunctionValues done in .19999999998354667E-005 cpu secs, .19073486328125000E-005 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse done in 1.9416529999999999 cpu secs, .60478305816650391 wall clock secs.
TIMER: GP sparsify done in 1.9490180000000001 cpu secs, .61529493331909180 wall clock secs.
libAtoms::Finalise: 07/10/2019 17:17:00
libAtoms::Finalise: Bye-Bye!
We can load and use the fitted potential:
[37]:
gap2b = Potential(param_filename='/tmp/gap_2b.xml')
We can calculate the potential energies using the new potential and compare them with the TB DFTB ones
[38]:
qm_energies = [at.info['energy'] for at in db]
gap2b_energies = []
for dba in db:
a = dba.copy()
a.set_calculator(gap2b)
gap2b_energies.append(a.get_potential_energy())
[39]:
Natoms = len(db[0])
plt.scatter(np.array(qm_energies)/Natoms, np.array(gap2b_energies)/Natoms)
plt.plot([E0-5.0, E0-5.0+0.05], [E0-5.0, E0-5.0+0.05], "k-")
plt.show()
We can also calculate the RMSE (in meV) as follows:
[40]:
np.sqrt(sum((np.array(qm_energies)/Natoms - np.array(gap2b_energies)/Natoms)**2)/len(gap2b_energies))
[40]:
0.035992967464800296
We can generate dimers for visualising the pairwise potential:
[41]:
dimers = [Atoms("2Si", positions=[[0,0,0], [x, 0,0]]) for x in np.linspace(1.6,6,100)]
[42]:
dimer_curve = []
for dim in dimers:
dim.set_calculator(gap2b)
dimer_curve.append(dim.get_potential_energy())
[43]:
plt.plot([dim.positions[1,0] for dim in dimers], np.array(dimer_curve)/2.0)
plt.show()
Fitting the 2 and 3-body potential
[44]:
!gap_fit at_file=/tmp/atoms_db.xyz \
gap={distance_Nb order=2 \
cutoff=5.0 \
covariance_type=ARD_SE \
theta_uniform=1.0 \
n_sparse=15 \
delta=1.0:\
distance_Nb order=3 \
cutoff=4.0 \
covariance_type=ARD_SE \
theta_uniform=1.0 \
n_sparse=50 \
delta=0.004} \
e0=-29.716948405885105 \
default_sigma={0.005 0.5 0.0 0.0} \
do_copy_at_file=F sparse_separate_file=F \
gp_file=/tmp/gap_3b.xml
libAtoms::Hello World: 07/10/2019 17:17:09
libAtoms::Hello World: git version git@github.com:/libAtoms/QUIP,d204dca49-dirty
libAtoms::Hello World: QUIP_ARCH darwin_x86_64_gfortran_openmp
libAtoms::Hello World: compiled on Aug 7 2019 at 14:13:45
libAtoms::Hello World: OpenMP parallelisation with 4 threads
WARNING: libAtoms::Hello World: environment variable OMP_STACKSIZE not set explicitly. The default value - system and compiler dependent - may be too small for some applications.
libAtoms::Hello World: Random Seed = 62229764
libAtoms::Hello World: global verbosity = 0
Calls to system_timer will do nothing by default
================================ Input parameters ==============================
at_file = /tmp/atoms_db.xyz
gap = "distance_Nb order=2 cutoff=5.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=15 delta=1.0: distance_Nb order=3 cutoff=4.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=50 delta=0.004"
e0 = -29.716948405885105
local_property0 = 0.0
e0_offset = 0.0
e0_method = isolated
default_sigma = "0.005 0.5 0.0 0.0"
sparse_jitter = 1.0e-10
hessian_delta = 1.0e-2
core_param_file = quip_params.xml
core_ip_args =
energy_parameter_name = energy
local_property_parameter_name = local_property
force_parameter_name = force
virial_parameter_name = virial
hessian_parameter_name = hessian
config_type_parameter_name = config_type
sigma_parameter_name = sigma
config_type_sigma =
sigma_per_atom = T
do_copy_at_file = F
sparse_separate_file = F
sparse_use_actual_gpcov = F
gp_file = /tmp/gap_3b.xml
verbosity = NORMAL
rnd_seed = -1
do_ip_timing = F
template_file = template.xyz
sparsify_only_no_fit = F
======================================== ======================================
============== Gaussian Approximation Potentials - Database fitting ============
Initial parsing of command line arguments finished.
Found 2 GAPs.
Descriptors have been parsed
XYZ file read
Unchanged GAP: {distance_Nb order=2 cutoff=5.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=15 delta=1.0}
Unchanged GAP: { distance_Nb order=3 cutoff=4.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=50 delta=0.004}
Multispecies support added where requested
===================== Report on number of descriptors found ====================
---------------------------------------------------------------------
Descriptor: distance_Nb order=2 cutoff=5.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=15 delta=1.0
Number of descriptors: 35312
Number of partial derivatives of descriptors: 0
---------------------------------------------------------------------
Descriptor: distance_Nb order=3 cutoff=4.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=50 delta=0.004
Number of descriptors: 150164
Number of partial derivatives of descriptors: 0
======================================== ======================================
========== Report on number of target properties found in training XYZ: ========
Number of target energies (property name: energy) found: 100
Number of target local_properties (property name: local_property) found: 0
Number of target forces (property name: force) found: 0
Number of target virials (property name: virial) found: 0
Number of target Hessian eigenvalues (property name: hessian) found: 0
================================= End of report ================================
===== Report on per-configuration/per-atom sigma (error parameter) settings ====
Number of per-configuration setting of energy_sigma found: 0
Number of per-configuration setting of force_sigma found: 0
Number of per-configuration setting of virial_sigma found: 0
Number of per-configuration setting of hessian_sigma found: 0
Number of per-atom setting of force_atom_sigma found: 0
================================= End of report ================================
Cartesian coordinates transformed to descriptors
Started sparse covariance matrix calculation of coordinate 1
Finished sparse covariance matrix calculation of coordinate 1
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1_sparse done in 1.9191969999999996 cpu secs, .61768078804016113 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1 done in 1.9192260000000001 cpu secs, .61770915985107422 wall clock secs.
Started sparse covariance matrix calculation of coordinate 2
Finished sparse covariance matrix calculation of coordinate 2
TIMER: gpFull_covarianceMatrix_sparse_Coordinate2_sparse done in 53.109310000000001 cpu secs, 15.321249008178711 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate2 done in 53.109337000000004 cpu secs, 15.321274995803833 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_LinearAlgebra done in .18150000000005662E-002 cpu secs, .74031352996826172E-002 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_FunctionValues done in .40000000041118255E-005 cpu secs, .28610229492187500E-005 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse done in 55.030578999999996 cpu secs, 15.946585178375244 wall clock secs.
TIMER: GP sparsify done in 55.042788999999999 cpu secs, 15.960463762283325 wall clock secs.
libAtoms::Finalise: 07/10/2019 17:17:29
libAtoms::Finalise: Bye-Bye!
We can load and use the fitted potential:
[45]:
gap3b = Potential(param_filename='/tmp/gap_3b.xml')
gap3b_energies = []
for at in db:
a = at.copy()
a.set_calculator(gap3b)
gap3b_energies.append(a.get_potential_energy())
We can also calculate the RMSE (in meV) as follows:
[46]:
np.sqrt(sum((np.array(qm_energies)/Natoms - np.array(gap3b_energies)/Natoms)**2)/len(gap3b_energies))
[46]:
0.010408552706046549
[47]:
plt.scatter(np.array(qm_energies)/Natoms, np.array(gap3b_energies)/Natoms)
plt.show()
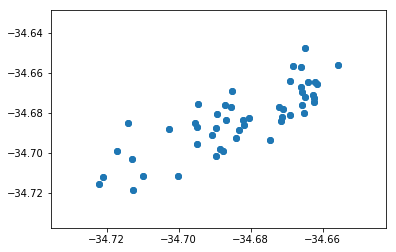
We can generate dimers for visualising the pairwise potential:
[48]:
dimer_curve = []
for dim in dimers:
dim.set_calculator(gap3b)
dimer_curve.append(dim.get_potential_energy())
plt.plot([dim.positions[1,0] for dim in dimers], np.array(dimer_curve)/2.0)
plt.show()
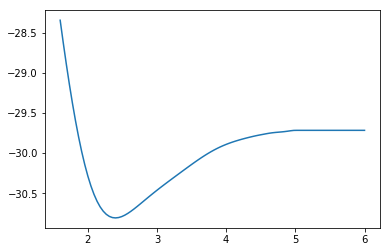
Fitting the 2 and 3-body and SOAP potential
We will describe that environment using the SOAP descriptor and compare them using the SOAP kernel. Note right away that the quantum mechanically computed energies are not fully determined by the near-environment of atoms, so this is an early indication that we will be making use of the “noise” interpretation of the \(\lambda\) regularization parameter: we don’t expect (and don’t want) our fitted function to precisely go through each datapoint.
The SOAP descriptor of an atomic environment is based on a spherical harmonic expansion of the neighbour density, and truncating this expansion at some maximum numer of radial (n_max) and angular (l_max) indices gives rise to some parameters. We also need to give the cutoff within which we consider the neighbour environment.
Writing the descriptor vector as \(p_{ss'nn'l}\), where \(s\) and \(s'\) are indices that run over the different atomic species in the atom’s environment, \(n\) and \(n'\) are radial and \(l\) is an angular index, the kernel between two atomic environments is
The factor of \(\delta^2\) allows the setting of the scale of fitted function, relative to the error specification \(\lambda\).
[49]:
!gap_fit at_file=/tmp/atoms_db.xyz \
gap={distance_Nb order=2 \
cutoff=5.0 \
covariance_type=ARD_SE \
theta_uniform=1.0 \
n_sparse=15 \
delta=1.0:\
distance_Nb order=3 \
cutoff=4.0 \
covariance_type=ARD_SE \
theta_uniform=1.0 \
n_sparse=50 \
delta=0.004:\
soap cutoff=4.0 \
covariance_type=dot_product \
zeta=2 \
delta=0.016 \
atom_sigma=0.7 \
l_max=6 \
n_max=6 \
n_sparse=200 \
sparse_method=cur_points} \
e0=-29.716948405885105 \
default_sigma={0.001 0.5 0.0 0.0} \
do_copy_at_file=F sparse_separate_file=F \
gp_file=/tmp/gap_2b3bsoap.xml 2>&1 | grep -v FoX
libAtoms::Hello World: 07/10/2019 17:17:32
libAtoms::Hello World: git version git@github.com:/libAtoms/QUIP,d204dca49-dirty
libAtoms::Hello World: QUIP_ARCH darwin_x86_64_gfortran_openmp
libAtoms::Hello World: compiled on Aug 7 2019 at 14:13:45
libAtoms::Hello World: OpenMP parallelisation with 4 threads
WARNING: libAtoms::Hello World: environment variable OMP_STACKSIZE not set explicitly. The default value - system and compiler dependent - may be too small for some applications.
libAtoms::Hello World: Random Seed = 62252544
libAtoms::Hello World: global verbosity = 0
Calls to system_timer will do nothing by default
================================ Input parameters ==============================
at_file = /tmp/atoms_db.xyz
gap = "distance_Nb order=2 cutoff=5.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=15 delta=1.0: distance_Nb order=3 cutoff=4.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=50 delta=0.004: soap cutoff=4.0 covariance_type=dot_product zeta=2 delta=0.016 atom_sigma=0.7 l_max=6 n_max=6 n_sparse=200 sparse_method=cur_points"
e0 = -29.716948405885105
local_property0 = 0.0
e0_offset = 0.0
e0_method = isolated
default_sigma = "0.001 0.5 0.0 0.0"
sparse_jitter = 1.0e-10
hessian_delta = 1.0e-2
core_param_file = quip_params.xml
core_ip_args =
energy_parameter_name = energy
local_property_parameter_name = local_property
force_parameter_name = force
virial_parameter_name = virial
hessian_parameter_name = hessian
config_type_parameter_name = config_type
sigma_parameter_name = sigma
config_type_sigma =
sigma_per_atom = T
do_copy_at_file = F
sparse_separate_file = F
sparse_use_actual_gpcov = F
gp_file = /tmp/gap_2b3bsoap.xml
verbosity = NORMAL
rnd_seed = -1
do_ip_timing = F
template_file = template.xyz
sparsify_only_no_fit = F
======================================== ======================================
============== Gaussian Approximation Potentials - Database fitting ============
Initial parsing of command line arguments finished.
Found 3 GAPs.
Descriptors have been parsed
XYZ file read
Unchanged GAP: {distance_Nb order=2 cutoff=5.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=15 delta=1.0}
Unchanged GAP: { distance_Nb order=3 cutoff=4.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=50 delta=0.004}
Unchanged GAP: { soap cutoff=4.0 covariance_type=dot_product zeta=2 delta=0.016 atom_sigma=0.7 l_max=6 n_max=6 n_sparse=200 sparse_method=cur_points}
Multispecies support added where requested
===================== Report on number of descriptors found ====================
---------------------------------------------------------------------
Descriptor: distance_Nb order=2 cutoff=5.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=15 delta=1.0
Number of descriptors: 35312
Number of partial derivatives of descriptors: 0
---------------------------------------------------------------------
Descriptor: distance_Nb order=3 cutoff=4.0 covariance_type=ARD_SE theta_uniform=1.0 n_sparse=50 delta=0.004
Number of descriptors: 150164
Number of partial derivatives of descriptors: 0
---------------------------------------------------------------------
Descriptor: soap cutoff=4.0 covariance_type=dot_product zeta=2 delta=0.016 atom_sigma=0.7 l_max=6 n_max=6 n_sparse=200 sparse_method=cur_points
Number of descriptors: 3200
Number of partial derivatives of descriptors: 0
======================================== ======================================
========== Report on number of target properties found in training XYZ: ========
Number of target energies (property name: energy) found: 100
Number of target local_properties (property name: local_property) found: 0
Number of target forces (property name: force) found: 0
Number of target virials (property name: virial) found: 0
Number of target Hessian eigenvalues (property name: hessian) found: 0
================================= End of report ================================
===== Report on per-configuration/per-atom sigma (error parameter) settings ====
Number of per-configuration setting of energy_sigma found: 0
Number of per-configuration setting of force_sigma found: 0
Number of per-configuration setting of virial_sigma found: 0
Number of per-configuration setting of hessian_sigma found: 0
Number of per-atom setting of force_atom_sigma found: 0
================================= End of report ================================
Started CUR decomposition
cur_decomposition: iteration: 1, error: .34329063034235423E-015
Finished CUR decomposition
Cartesian coordinates transformed to descriptors
Started sparse covariance matrix calculation of coordinate 1
Finished sparse covariance matrix calculation of coordinate 1
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1_sparse done in 1.9762869999999992 cpu secs, .54421901702880859 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate1 done in 1.9763460000000004 cpu secs, .54428696632385254 wall clock secs.
Started sparse covariance matrix calculation of coordinate 2
Finished sparse covariance matrix calculation of coordinate 2
TIMER: gpFull_covarianceMatrix_sparse_Coordinate2_sparse done in 52.650658000000000 cpu secs, 14.171960830688477 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate2 done in 52.650689000000000 cpu secs, 14.171991109848022 wall clock secs.
Started sparse covariance matrix calculation of coordinate 3
Covariance matrix 100% |********************| 0.3 / 0.3 s
Finished sparse covariance matrix calculation of coordinate 3
TIMER: gpFull_covarianceMatrix_sparse_Coordinate3_sparse done in .69204299999999819 cpu secs, .25782394409179688 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_Coordinate3 done in .69208299999999667 cpu secs, .25786900520324707 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_LinearAlgebra done in .10295999999999594 cpu secs, .42098045349121094E-001 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse_FunctionValues done in .20000000020559128E-005 cpu secs, .21457672119140625E-005 wall clock secs.
TIMER: gpFull_covarianceMatrix_sparse done in 55.422404999999998 cpu secs, 15.016569852828979 wall clock secs.
TIMER: GP sparsify done in 55.621861000000003 cpu secs, 15.223239183425903 wall clock secs.
libAtoms::Finalise: 07/10/2019 17:17:51
libAtoms::Finalise: Bye-Bye!
We can load and use the fitted potential:
[50]:
gap_soap = Potential(param_filename='/tmp/gap_2b3bsoap.xml')
gap_energies = []
for at in db:
a = at.copy()
a.set_calculator(gap_soap)
gap_energies.append(a.get_potential_energy())
We can also calculate the RMSE (in meV) as follows:
[51]:
np.sqrt(sum((np.array(qm_energies)/Natoms - np.array(gap_energies)/Natoms)**2)/len(gap_energies))
[51]:
0.004254157149981674
[52]:
plt.scatter(np.array(qm_energies)/Natoms, np.array(gap_energies)/Natoms)
plt.show()
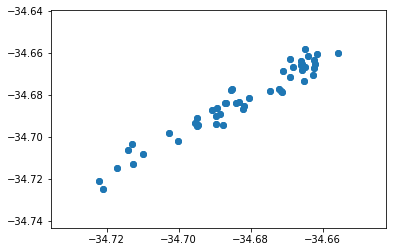
Using GAP for MD
We can use now use the fitted potential to do MD or geometry relaxation:
[53]:
#attach tight binding calculator
atoms.set_calculator(gap_soap)
# Thermalize atoms
# MaxwellBoltzmannDistribution(atoms, 2.0* T * units.kB)
# dynamics = VelocityVerlet(atoms, timestep)
dynamics = Langevin(atoms, timestep, T * units.kB, 0.002)
dynamics.attach(print_status, interval=10)
dynamics.attach(print_energy, interval=10)
Dynamics:
[54]:
dynamics.run(steps=100)
Step = 0, time = 0.0 [fs], T = 1294.479828341841 [K]
Energy per atom: Epot = -34.698eV Ekin = 0.167eV (T=1294K) Etot = -34.531eV
Step = 10, time = 10.0 [fs], T = 1189.7904138587505 [K]
Energy per atom: Epot = -34.685eV Ekin = 0.154eV (T=1190K) Etot = -34.531eV
Step = 20, time = 20.0 [fs], T = 1264.3379093179376 [K]
Energy per atom: Epot = -34.696eV Ekin = 0.163eV (T=1264K) Etot = -34.533eV
Step = 30, time = 30.0 [fs], T = 1457.3725237044011 [K]
Energy per atom: Epot = -34.721eV Ekin = 0.188eV (T=1457K) Etot = -34.533eV
Step = 40, time = 40.0 [fs], T = 1388.8679832826024 [K]
Energy per atom: Epot = -34.715eV Ekin = 0.180eV (T=1389K) Etot = -34.536eV
Step = 50, time = 50.00000000000001 [fs], T = 1561.736378264197 [K]
Energy per atom: Epot = -34.738eV Ekin = 0.202eV (T=1562K) Etot = -34.536eV
Step = 60, time = 60.0 [fs], T = 2028.6943305981565 [K]
Energy per atom: Epot = -34.800eV Ekin = 0.262eV (T=2029K) Etot = -34.538eV
Step = 70, time = 70.0 [fs], T = 1568.5562709553685 [K]
Energy per atom: Epot = -34.737eV Ekin = 0.203eV (T=1569K) Etot = -34.535eV
Step = 80, time = 80.0 [fs], T = 1886.3013570587461 [K]
Energy per atom: Epot = -34.779eV Ekin = 0.244eV (T=1886K) Etot = -34.535eV
Step = 90, time = 90.0 [fs], T = 1928.3315444589239 [K]
Energy per atom: Epot = -34.787eV Ekin = 0.249eV (T=1928K) Etot = -34.537eV
Step = 100, time = 100.00000000000001 [fs], T = 1727.5769477863184 [K]
Energy per atom: Epot = -34.761eV Ekin = 0.223eV (T=1728K) Etot = -34.538eV
[54]:
True
Geometry optimisation:
[55]:
optatoms = db[0].copy()
optatoms.set_calculator(gap_soap)
opt = PreconLBFGS(optatoms, precon=Exp(3.0))
[56]:
opt.run(fmax=0.01)
PreconLBFGS: 0 17:18:02 -1109.683388 4.9234
estimate_mu(): mu=1.4377293786564451, mu_c=1.0
PreconLBFGS: 1 17:18:02 -1110.535572 4.5917
PreconLBFGS: 2 17:18:02 -1111.272884 4.5079
PreconLBFGS: 3 17:18:02 -1111.957952 4.3767
PreconLBFGS: 4 17:18:02 -1112.578689 4.3500
PreconLBFGS: 5 17:18:02 -1113.225669 4.3169
PreconLBFGS: 6 17:18:02 -1113.863449 4.2918
PreconLBFGS: 7 17:18:03 -1114.471548 4.2458
PreconLBFGS: 8 17:18:03 -1115.032581 4.1511
PreconLBFGS: 9 17:18:03 -1115.574963 4.0274
PreconLBFGS: 10 17:18:03 -1116.119287 3.8739
PreconLBFGS: 11 17:18:03 -1116.680878 3.6919
PreconLBFGS: 12 17:18:03 -1117.209393 3.5311
PreconLBFGS: 13 17:18:03 -1117.716249 3.3508
PreconLBFGS: 14 17:18:03 -1118.160184 3.2262
PreconLBFGS: 15 17:18:03 -1118.601303 3.0357
PreconLBFGS: 16 17:18:03 -1118.990945 2.9062
PreconLBFGS: 17 17:18:04 -1119.349906 2.8058
PreconLBFGS: 18 17:18:04 -1119.678573 2.7172
PreconLBFGS: 19 17:18:04 -1120.006870 2.6478
PreconLBFGS: 20 17:18:04 -1120.342863 2.5801
PreconLBFGS: 21 17:18:04 -1120.661850 2.5140
PreconLBFGS: 22 17:18:04 -1120.989318 2.4015
PreconLBFGS: 23 17:18:04 -1121.302870 2.4258
PreconLBFGS: 24 17:18:04 -1121.594728 2.5018
PreconLBFGS: 25 17:18:04 -1121.864728 2.5801
PreconLBFGS: 26 17:18:04 -1122.124053 2.5994
PreconLBFGS: 27 17:18:04 -1122.396679 2.6003
PreconLBFGS: 28 17:18:05 -1122.669355 2.5872
PreconLBFGS: 29 17:18:05 -1122.963355 2.4957
PreconLBFGS: 30 17:18:05 -1123.269795 2.3171
PreconLBFGS: 31 17:18:05 -1123.603594 1.9910
PreconLBFGS: 32 17:18:05 -1123.937762 1.5578
PreconLBFGS: 33 17:18:05 -1124.234675 1.3468
PreconLBFGS: 34 17:18:05 -1124.513655 1.1899
PreconLBFGS: 35 17:18:05 -1124.758552 1.2400
PreconLBFGS: 36 17:18:05 -1124.966145 1.2919
PreconLBFGS: 37 17:18:05 -1125.150890 1.3222
PreconLBFGS: 38 17:18:06 -1125.313512 1.2540
PreconLBFGS: 39 17:18:06 -1125.484647 1.2145
PreconLBFGS: 40 17:18:06 -1125.645388 1.1570
PreconLBFGS: 41 17:18:06 -1125.802081 1.1058
PreconLBFGS: 42 17:18:06 -1125.944609 1.0548
PreconLBFGS: 43 17:18:06 -1126.066383 1.0593
PreconLBFGS: 44 17:18:06 -1126.162365 1.0664
PreconLBFGS: 45 17:18:06 -1126.246577 1.0189
PreconLBFGS: 46 17:18:06 -1126.330379 1.0728
PreconLBFGS: 47 17:18:06 -1126.410825 1.1446
PreconLBFGS: 48 17:18:07 -1126.488125 1.2435
PreconLBFGS: 49 17:18:07 -1126.560200 1.3443
PreconLBFGS: 50 17:18:07 -1126.626230 1.4196
PreconLBFGS: 51 17:18:07 -1126.686193 1.3669
PreconLBFGS: 52 17:18:07 -1126.742823 1.3389
PreconLBFGS: 53 17:18:07 -1126.795906 1.2727
PreconLBFGS: 54 17:18:07 -1126.845887 1.2176
PreconLBFGS: 55 17:18:07 -1126.893799 1.1407
PreconLBFGS: 56 17:18:07 -1126.940278 1.0719
PreconLBFGS: 57 17:18:07 -1126.982981 0.9973
PreconLBFGS: 58 17:18:08 -1127.029085 0.9026
PreconLBFGS: 59 17:18:08 -1127.070015 0.8517
PreconLBFGS: 60 17:18:08 -1127.116317 0.8929
PreconLBFGS: 61 17:18:08 -1127.161266 0.9901
PreconLBFGS: 62 17:18:08 -1127.202419 1.0645
PreconLBFGS: 63 17:18:08 -1127.241068 1.0903
PreconLBFGS: 64 17:18:08 -1127.282508 1.0586
PreconLBFGS: 65 17:18:08 -1127.347191 0.8213
PreconLBFGS: 66 17:18:08 -1127.405873 0.7884
PreconLBFGS: 67 17:18:09 -1127.448727 0.7998
PreconLBFGS: 68 17:18:09 -1127.486176 0.7780
PreconLBFGS: 69 17:18:09 -1127.525671 0.7810
PreconLBFGS: 70 17:18:09 -1127.558559 0.8359
PreconLBFGS: 71 17:18:09 -1127.598349 0.8139
PreconLBFGS: 72 17:18:09 -1127.634101 0.8673
PreconLBFGS: 73 17:18:09 -1127.662738 0.8824
PreconLBFGS: 74 17:18:09 -1127.687485 0.9169
PreconLBFGS: 75 17:18:09 -1127.698444 1.0778
Passed direction which is not downhill. Aborting...
PreconLBFGS: 76 17:18:09 -1127.698444 1.0778
PreconLBFGS: 77 17:18:10 -1127.719816 1.0899
/Users/jameskermode/.pyenv/versions/3.6.6/Python.framework/Versions/3.6/lib/python3.6/site-packages/ase/optimize/precon/lbfgs.py:352: UserWarning: Armijo linesearch failed, resetting Hessian and trying again
'Armijo linesearch failed, resetting Hessian and '
PreconLBFGS: 78 17:18:10 -1127.761700 1.1322
PreconLBFGS: 79 17:18:10 -1127.844255 1.2291
PreconLBFGS: 80 17:18:10 -1127.969036 1.3938
PreconLBFGS: 81 17:18:10 -1128.081393 1.4121
PreconLBFGS: 82 17:18:10 -1128.181345 1.3558
PreconLBFGS: 83 17:18:10 -1128.270254 1.3927
PreconLBFGS: 84 17:18:10 -1128.348531 1.5283
PreconLBFGS: 85 17:18:10 -1128.426889 1.7799
PreconLBFGS: 86 17:18:11 -1128.445736 1.8243
Passed direction which is not downhill. Aborting...
PreconLBFGS: 87 17:18:11 -1128.445736 1.8243
PreconLBFGS: 88 17:18:11 -1128.482902 1.9309
PreconLBFGS: 89 17:18:11 -1128.559965 2.2312
Passed direction which is not downhill. Aborting...
PreconLBFGS: 90 17:18:11 -1128.559965 2.2312
PreconLBFGS: 91 17:18:11 -1128.670491 2.4921
Passed direction which is not downhill. Aborting...
PreconLBFGS: 92 17:18:11 -1128.670491 2.4921
PreconLBFGS: 93 17:18:11 -1128.782318 2.3591
PreconLBFGS: 94 17:18:11 -1128.894364 2.1521
PreconLBFGS: 95 17:18:11 -1129.001220 2.2479
PreconLBFGS: 96 17:18:11 -1129.106928 2.3316
Passed direction which is not downhill. Aborting...
PreconLBFGS: 97 17:18:11 -1129.106928 2.3316
PreconLBFGS: 98 17:18:11 -1129.220418 2.3091
PreconLBFGS: 99 17:18:12 -1129.331678 2.2090
PreconLBFGS: 100 17:18:12 -1129.443932 2.0163
PreconLBFGS: 101 17:18:12 -1129.551115 1.6623
PreconLBFGS: 102 17:18:12 -1129.652255 1.2197
PreconLBFGS: 103 17:18:12 -1129.733200 0.8105
PreconLBFGS: 104 17:18:12 -1129.793145 0.6091
PreconLBFGS: 105 17:18:12 -1129.830076 0.5602
PreconLBFGS: 106 17:18:12 -1129.844974 0.3316
PreconLBFGS: 107 17:18:12 -1129.853075 0.4243
PreconLBFGS: 108 17:18:13 -1129.857980 0.2636
PreconLBFGS: 109 17:18:13 -1129.861664 0.1665
PreconLBFGS: 110 17:18:13 -1129.863643 0.1211
PreconLBFGS: 111 17:18:13 -1129.864671 0.0906
PreconLBFGS: 112 17:18:13 -1129.865139 0.0449
PreconLBFGS: 113 17:18:13 -1129.865458 0.0324
PreconLBFGS: 114 17:18:14 -1129.865586 0.0232
PreconLBFGS: 115 17:18:14 -1129.865642 0.0139
PreconLBFGS: 116 17:18:14 -1129.865663 0.0100
PreconLBFGS: 117 17:18:14 -1129.865673 0.0062
[56]:
True
Let’s visualise the relaxed structure:
[57]:
view = ViewStructure(optatoms, (2,2,1))
view.camera= 'orthographic'
view